Abstract
The SARS-CoV-2 pandemic has seen a notable global reduction in influenza cases of both influenza A and B viruses. In particular, the B/Yamagata lineage has not been isolated from April 2020 to August 2021, suggesting that this influenza lineage may have become extinct, which may provide opportunities for improving availability and effectiveness of influenza vaccines.
Subject terms: Influenza virus, Viral epidemiology
Infection with influenza B virus (IBV) accounts for around a quarter of the annual influenza burden. Influenza vaccine composition is reconsidered bi-annually to account for continuous antigenic drift in the haemagglutinin protein. The co-circulation of two antigenically distinct lineages of IBV, referred to as B/Victoria/2/1987 and B/Yamagata/16/1988 lineages (hereafter B/Victoria and B/Yamagata), has driven the development and application of a quadrivalent influenza vaccine (QIV). With circulation of IBV only in humans and no established animal reservoir1, high vaccination coverage that induces broad immune protection provides a potential pathway to eradication of IBV. The considerable reductions of influenza circulation globally due to COVID-19 may have already precipitated the extinction of the B/Yamagata lineage.
Global disappearance of B/Yamagata viruses during the COVID-19 pandemic
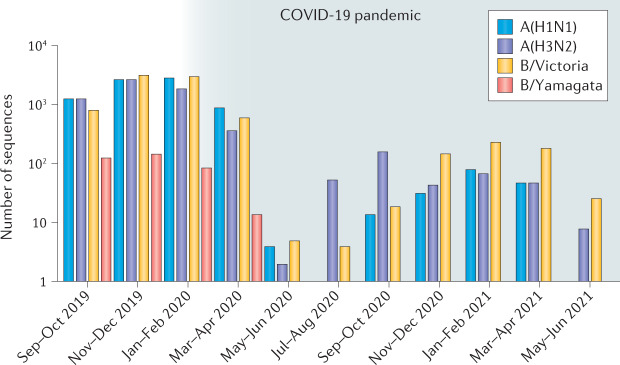
Influenza virus detections dropped dramatically from April 2020 (Fig. 1), with a ~99% reduction compared with previous years despite roughly similar levels of testing2. Behavioural changes (social distancing, mask wearing and hygiene measures) and travel and movement restrictions are thought to be the major factors driving the reduction in influenza incidence, which was also observed for other common respiratory infections such as respiratory syncytial virus3. The genetic diversity of influenza viruses has also dramatically diminished (Supplementary Fig. 1). The number of clades detected has fallen for both influenza A(H3N2) and A(H1N1) subtypes, and, similarly, diversity has diminished for the B/Victoria lineage. Although most IBV cases are not routinely subtyped into the two lineages, only very few B/Yamagata cases (none with successful virus isolation or sequencing) have been reported globally in 2021 (31 cases in FluNet as of 27 July 2021; Supplementary Table 1), and, since March 2020, no B/Yamagata viruses have been isolated or sequenced.
Fig. 1. Circulation of influenza viruses before and during the COVID-19 pandemic.
Global influenza virus sequences from the GISAID database with collection dates from September 2019 to 6 August 2021 illustrate a reduction of influenza virus detection and potential extinction of B/Yamagata during the COVID-19 pandemic (shaded).
Has the B/Yamagata lineage gone for good, or merely gone into hiding? While the global dominance of the B/Victoria and B/Yamagata lineages seems to alternate in recent years (Supplementary Fig. 2), IBV lineages have been known to periodically enter a state of ‘dormancy’ for long time intervals. For example, during the 1990s the B/Victoria lineage was mostly undetected globally, except for an outbreak in Asia, but subsequently dominated global IBV isolates in the early 2000s4.
Epidemiology and evolution of B/Yamagata viruses
The stark reduction in global prevalence of B/Yamagata viruses compared with the other lineages may indicate an inherent vulnerability of this lineage. Indeed, B/Yamagata viruses have a lower effective reproductive number than B/Victoria viruses, and B/Yamagata epidemics have a slower initial growth phase with shorter transmission chains than B/Victoria epidemics5. This may make B/Yamagata more vulnerable to breakdowns in onward transmission, especially in the context of social distancing and movement restrictions. Furthermore, although multiple B/Yamagata clades can co-circulate for extended periods, previously long-lived clades went extinct6,7. Although the precise factors that drive B/Yamagata clades into extinction are unknown, the frequency of such extinctions suggests an intrinsic volatility in global B/Yamagata circulation. Lastly, as B/Victoria prevalence has been increasing since 2019, B/Yamagata may have already been at a low prevalence cycle at the beginning of the pandemic.
The global circulation patterns of influenza viruses have been associated with age-specific patterns of infection, whereby infection of adults, who are more likely to travel than children, correlates with greater global spread8. School-aged children and adolescents are more frequently infected with B/Victoria viruses, whereas adults over the age of 25 years are more frequently infected with B/Yamagata viruses5,7. Consistently, in 2008–2019, B/Yamagata viruses demonstrated greater global movement than B/Victoria viruses7. The reduction in global mobility due to the COVID-19 pandemic may have contributed to low distribution and seeding of B/Yamagata. Overall, the temporal and intrinsic epidemiology of B/Yamagata viruses may have rendered them particularly vulnerable to this effect.
From 2017, circulating B/Yamagata viruses have all been from a single lineage (clade 3A), whereas several lineages existed before. Although this clade has diversified at the genomic level, and even acquired amino acid substitution in haemagglutinin, these have not resulted in significant antigenic changes to necessitate updating the B/Yamagata vaccine component since 2015 (refs6,7). Therefore, although B/Yamagata viruses have continued to evolve genetically, they remain antigenically conserved in the haemagglutinin protein in recent years. Thus, the onset of the COVID-19 pandemic coinciding with a period of low incidence and low antigenic diversity, coupled with prolonged use of a well-matched influenza vaccine, may have enabled massive suppression of B/Yamagata during the pandemic. However, as sampling and sequencing is not comprehensive, it is difficult to distinguish with certainty between lack of detection and true extinction.
Implications for vaccines
The potential extinction of one of the circulating seasonal IBV lineages creates a dilemma for the vaccine strain recommendation, with two possible alternative strategies. First, returning to vaccines with two IAV strains and a B/Victoria strain could increase the available doses for global distribution (from ~500 million QIV doses to ~700 million doses of a trivalent vaccine9). The second possibility is including two A(H3N2) candidate viruses in a QIV, as the divergence and co-circulation of A(H3N2) sub-clades often makes vaccine strain selection challenging. The inclusion of two A(H3N2) viruses should give greater antigenic coverage, in principle, although direct data for this are currently lacking and clinical trials would be needed with substantial cost. Furthermore, the inclusion of two A(H3N2) strains will be challenging with respect to the current registered potency assay, as the single radial immunodiffusion assay uses polyclonal sera, which may not distinguish between alternate A(H3N2) virus clades. Thus, alternative potency assays that use strain-specific reagents will be required.
Summary
Epidemiological and evolutionary characteristics of B/Yamagata viruses, combined with suppressive conditions of the COVID-19 pandemic, may have facilitated strong suppression of global B/Yamagata circulation and the potential extinction of this lineage. While time will tell whether the B/Yamagata lineage is gone for good, elimination of one of the four current vaccine targets would have favourable implications for annual influenza vaccine reformulation, with opportunities for rational rethinking of optimal strategies to further reduce global influenza burden.
Supplementary information
Acknowledgements
The authors thank Kirk Truong and Lynda Allan for help with the analysis of data included. M.K., A.K.W. and S.J.K are supported by National Health and Medical Research Council (NHMRC) fellowships.
Competing interests
K.L. and S.R. are employees of Seqirus, an influenza vaccine manufacturer. The other authors declare no competing interests.
Footnotes
Related links
FluNet: https://www.who.int/tools/flunet
Supplementary information
The online version contains supplementary material available at 10.1038/s41579-021-00642-4.
References
- 1.Koutsakos M, Nguyen THO, Barclay WS, Kedzierska K. Knowns and unknowns of influenza B viruses. Future Microbiol. 2016;11:119–135. doi: 10.2217/fmb.15.120. [DOI] [PubMed] [Google Scholar]
- 2.Laurie KL, Rockman S. Which influenza viruses will emerge following the SARS-CoV-2 pandemic? Influenza Other Respir. Viruses. 2021 doi: 10.1111/irv.12866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang QS, et al. Impact of the COVID-19 nonpharmaceutical interventions on influenza and other respiratory viral infections in New Zealand. Nat. Commun. 2021;12:1001. doi: 10.1038/s41467-021-21157-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen R, Holmes EC. The evolutionary dynamics of human influenza B virus. J. Mol. Evol. 2008;66:655–663. doi: 10.1007/s00239-008-9119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vijaykrishna D, et al. The contrasting phylodynamics of human influenza B viruses. Elife. 2015;4:e05055. doi: 10.7554/eLife.05055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langat P, et al. Genome-wide evolutionary dynamics of influenza B viruses on a global scale. PLoS Pathog. 2017;13:e1006749. doi: 10.1371/journal.ppat.1006749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Virk RK, et al. Divergent evolutionary trajectories of influenza B viruses underlie their contemporaneous epidemic activity. Proc. Natl Acad. Sci. USA. 2020;117:619–628. doi: 10.1073/pnas.1916585116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bedford T, et al. Global circulation patterns of seasonal influenza viruses vary with antigenic drift. Nature. 2015;523:217–220. doi: 10.1038/nature14460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palache A, et al. Vaccine complacency and dose distribution inequities limit the benefits of seasonal influenza vaccination, despite a positive trend in use. Vaccine. 2021;39:6081–6087. doi: 10.1016/j.vaccine.2021.08.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.