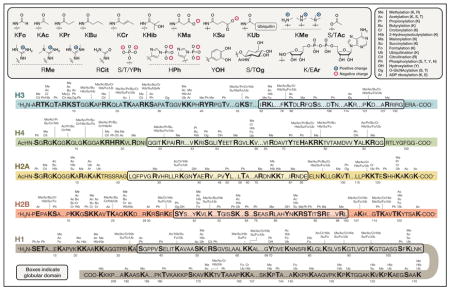
The histone proteins are decorated by a variety of protein posttranslational modifications (also called histone marks). Histone marks are critical to dynamic modulation of chromatin structure and function, contributing to the cellular gene expression program. In addition to the well-studied acetylation and methylation modifications, recent studies have revealed several new types of histone marks, including lysine propionylation, lysine butyrylation, lysine crotonylation, lysine 2-hydroxyisobutyrylation, lysine malonylation, and lysine succinylation. Preliminary studies on some of the new histone marks (e.g., crotonylation and 2-hydroxyisobutyrylation) suggest that their effects on chromatin function are distinct from those of lysine acetylation. Given that the newly discovered lysine acylation reactions likely use the corresponding acyl-CoA molecules as cofactors, it is proposed that histone acylations provide a link between cellular metabolism and epigenetic mechanisms.
This SnapShot summarizes the reported human, mouse, and rat histone marks, including recently identified lysine acylation marks.
Acknowledgments
This work was funded by institutional support from The Rockefeller University to C.D.A.; by NIH grants GM105933, CA160036, and RR020839 to Y.Z.; and by DP2OD007447 to B.A.G.
References
- Arnaudo AM, Garcia BA. Epigenet Chromatin. 2013;6:24. doi: 10.1186/1756-8935-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck HC, Nielsen EC, Matthiesen R, Jensen LH, Sehested M, Finn P, Grauslund M, Hansen AM, Jensen ON. Mol Cell Proteomics. 2006;5:1314–1325. doi: 10.1074/mcp.M600007-MCP200. [DOI] [PubMed] [Google Scholar]
- Chen Y, Sprung R, Tang Y, Ball H, Sangras B, Kim SC, Falck JR, Peng J, Gu W, Zhao Y. Mol Cell Proteomics. 2007;6:812–819. doi: 10.1074/mcp.M700021-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L, Peng C, Montellier E, Lu Z, Chen Y, Ishii H, Debernardi A, Buchou T, Rousseaux S, Jin F, et al. Nat Chem Biol. 2014;10:365–370. doi: 10.1038/nchembio.1497. [DOI] [PubMed] [Google Scholar]
- Fierz B, Muir TW. Nat Chem Biol. 2012;8:417–427. doi: 10.1038/nchembio.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al. Cell. 2011;146:1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Dai J, Dai L, Tan M, Cheng Z, Wu Y, Boeke JD, Zhao Y. Mol Cell Proteomics. 2012;11:100–107. doi: 10.1074/mcp.M111.015875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NL, Dimaggio PA, Garcia BA. Cell Mol Life Sci. 2010;67:3983–4000. doi: 10.1007/s00018-010-0475-7. [DOI] [PMC free article] [PubMed] [Google Scholar]


