Abstract
Esophageal adenocarcinoma (EAC) is a poor prognosis cancer type with rapidly rising incidence. Our understanding of genetic events that drive EAC development is limited, and there are few molecular biomarkers for prognostication or therapeutics. Using a cohort of 551 genomically characterized EACs with matched RNA-seq, we discover 77 EAC driver genes and 21 non-coding driver elements. We identify a mean of 4.4 driver events per tumor, derived more commonly from mutations rather than copy number alterations, and compare these mutations to the exome-wide mutational excess using dN/dS calculations. We observe mutual exclusivity or co-occurrence of events within and between a number of dysregulated EAC pathways, suggestive of strong functional relationships. Poor prognostic indicators (SMAD4, GATA4) are verified in independent cohorts with significant predictive value. Over 50% of EACs contain sensitizing events for CDK4/6 inhibitors, which are highly correlated with clinically relevant sensitivity in a panel EAC cell lines and organoids.
Esophageal cancer is the eighth most common form of cancer world-wide and the sixth most common cause of cancer related death1. Esophageal adenocarcinoma (EAC) is the predominant subtype in the west, and incidence has been rapidly rising2. EAC is a highly aggressive neoplasm, usually presenting at a late stage, and is generally resistant to chemotherapy, leading to five-year survival rates below 15%3. It is characterized by very high mutation rates in comparison to other cancer types4 but also, paradoxically, by a paucity of recurrently mutated genes. EAC displays dramatic chromosomal instability and thus may be classified as a C-type neoplasm, which may be driven mainly by structural variation rather than mutations5,6. Currently, our understanding of precisely which genetic events drive the development of EAC is limited, and consequentially there is a paucity of molecular biomarkers for prognosis or targeted therapeutics available.
Methods to differentiate driver mutations from passenger mutations use features associated with known drivers to detect regions of the genome in which mutations are enriched for these features7. The simplest of these features is the tendency of a mutation to co-occur with other mutations in the same gene at a high frequency, as detected by MutSigCV8. MutSigCV has identified 12 known cancer genes as EAC drivers (TP53, CDKN2A, SMAD4, ARID1A, ERBB2, KRAS, PIK3CA, SMARCA4, CTNNB1, ARID2, PBRM1 and FBXW7)6,9,10. The PCAWG ICGC analysis also identified a significantly mutated enhancer associated with TP53TG111. However, these analyses leave most EAC cases with only one known driver mutation, usually TP53. Equivalent analyses in other cancer types have identified three or four drivers per case12,13. Similarly, detection of copy number driver events in EAC has relied on identifying regions of the genome recurrently deleted or amplified, as detected by GISTIC9,14–17. However, GISTIC often identifies relatively large regions of the genome, with little indication of which specific gene-copy number aberrations (CNAs) may actually confer a selective advantage. There are also several non-selection based mechanisms that can cause recurrent CNAs, such as genomic fragile sites, which have not been well differentiated from selection-based CNAs18. Epigenetic events, for example methylation, may also be important sources of driver events in EAC but are much more difficult to assess formally for selection.
To address these issues, we accumulated a cohort of 551 genomically characterized EACs using our esophageal ICGC project, which have high quality clinical annotation, associated whole genome sequencing (WGS) and RNA-seq on cases with sufficient material. We augmented our ICGC WGS cohort with publicly available whole exome19 and whole genome sequencing20 data and applied a number of complementary driver detection methods to produce a comprehensive assessment of mutations and CNAs under selection in EAC. We use these events to define functional cell processes that have been selectively dysregulated in EAC and identify novel, verifiable and clinically relevant biomarkers for prognostication. Finally, we have used this compendium of EAC driver events to provide an evidence base for targeted therapeutics, which we have tested in vitro.
Results
A compendium of EAC driver events and their functional impact
In 551 EACs, we identified a total of 11,813,333 single nucleotide variants (SNVs) and small insertions or deletions (Indels), with a median of 6.4 such mutations/Mb (Supplementary Fig. 1), and 286,965 copy number aberrations (CNAs). We also identified 134,697 structural variants (SVs) in WGS cases. We use several complementary driver detection tools to detect driver-associated features in mutations and CNAs (Fig. 1a). Each tool underwent quality control to ensure reliability of results (see Methods). These features include highly recurrent mutations within a gene (dNdScv21, ActivedriverWGS22, MutSigCV28), high functional impact mutations within a gene (OncodriveFM23, ActivedriverWGS22), mutation clustering (OncodriveClust24, eDriver25 and eDriver3D26) and recurrent amplification or deletion of genes (GISTIC14) undergoing concurrent over or under-expression (see Methods) (Fig. 1a)7.
Figure 1. Detection of EAC driver genes.
a, Types of driver-associated features used to detect positive selection in mutations and copy number events with examples of genes containing such features. b, Coding driver genes identified and their driver-associated features. c, Non-coding driver elements detected and their element types.
These complementary methods produced highly significant agreement in calling EAC driver genes, particularly within the same feature-type (Supplementary Fig. 2), and on average more than half of the genes identified by one feature were also identified by other features (Fig. 1b). In total, 76 EAC driver genes were discovered, 71% of which have not been detected in EAC previously9,10,15–17,19 and 69% of which are known drivers in pan-cancer analyses21,27,28. To detect driver elements in the non-coding genome, we used ActiveDriverWGS22, a recently benchmarked29 method using both functional impact and recurrence to determine driver status (Fig. 1c and Supplementary Fig. 3). We discovered 21 non-coding driver elements using this method. We have recovered several known non-coding driver elements from the pan-cancer PCAWG analysis11, including the enhancer on chromosome 7 linked to TP53TG1 previously identified in EAC and the promoter/5’UTR regions of PTDSS1 and WRD74 found in other cancer types. We also identified novel non-coding cancer driver elements, including in the 5’UTR of MMP24 and promoters of two related histones (HIST1H2BO and HIST1H2AM).
EAC is notable among cancer types for harboring a high degree of chromosomal instability20. Using GISTIC, we identified 149 recurrently deleted or amplified loci across the genome (Fig. 2a, and Supplementary Tables 1 and 2). To determine which genes within these loci confer a selective advantage when they undergo CNAs, we use a subset of 116 cases with matched RNA-seq to detect genes in which homozygous deletion or amplification causes a significant under or over-expression, respectively (Supplementary Note and Supplementary Tables 3-6). The majority of genes in these regions showed no significant copy number associated expression change (74%), although work in larger cohorts suggests we may be underpowered to detect small expression changes30. We observed highly significant expression changes in 17 known cancer genes within GISTIC loci such as ERBB2, KRAS and SMAD4, which we designate high-confidence EAC drivers (see Methods). We also found five tumor suppressor genes where copy number loss was not necessarily associated with expression modulation but tightly associated with presence of mutations leading to LOH, for example ARID1A and CDH11.
Figure 2. Copy number variation under positive selection.
a, Recurrent copy number changes across the genome identified by GISTIC in 551 EACs. Frequency of different CNV types are indicated (dark blue, homozygous deletion; light blue, heterozygous deletion; dark red, extrachromosomal-like amplification; light red, amplification) as well as the position of CNV high confidence driver genes and candidate driver genes. The q value for expression correlation with amplification and homozygous deletion is shown for each gene within each amplification (wilcox test, one sided, expression compared above and below 90th percentile of pliody-adjusted CN) and deletion peak (wilcox test, one sided, expression compared between homozygous deleted and all other cases) respectively and occasions of significant association between LOH and mutation are indicated in green (fisher’s exact test, one sided). Benjamini & Hochberg false discovery correction was applied in each of these cases. Purple deletion peaks indicate fragile sites. b, Examples of extrachromosomal-like amplifications suggested by very high read support SVs at the boundaries of highly amplified regions produced from a single copy number step. In the first example two populations of extrachromosomal DNA are apparent, one amplifying only MYC and the second also incorporating ERBB2 from a different chromosome. In the second example an inversion has occurred before circularization and amplification around KRAS. c, Relationship between copy number and expression in copy number driver genes in RNA matched sub-cohort (n=116). A 2D kernel density estimation and a leoss regression curve with 95% CIs (grey) are shown to describe the data.
In a subset of GISTIC loci, we observed extremely high copy number amplification, commonly greater than 100 copies, and these events were highly enriched in recurrently amplified regions containing driver genes rather than those which appear to contain only passengers (ploidy adjusted copy number >10, two-sided Wilcoxon test, P = 4.97 x 10-8) (Supplementary Fig. 4). We use ploidy adjusted copy number to define amplifications as it produces superior correlation with expression data than absolute copy number alone. Ploidy of our samples varies from 1.4-6.2 (median 2.8), and hence ploidy adjusted copy number of >10 cut off translates into >14-62 absolute copies (on average 28 copies). To discern a mechanism for these ultra-high amplifications, we assessed structural variants (SVs) associated with these events. For many of these events, the extreme amplification was produced largely from a single copy number step, the edges of which were linked by structural variants with ultra-high read support. Two examples are shown in Figure 2b, and further randomly selected examples in Supplementary Figure 5. In the first example, circularization and amplification initially occurred around MYC but subsequently incorporated ERBB2 from an entirely different chromosome, and in the second, an inversion was followed by circularization and amplification of KRAS. Such a pattern of extrachromosomal amplification via double minutes has been previously noted in EAC20 and other neoplasms31, and hence we refer to this amplification class with ultra-high amplification (ploidy adjusted copy number >10) as ‘extrachromosomal-like’.
We found that extrachromosomal-like amplifications had extreme and highly penetrant effects on expression, while moderate amplification (ploidy adjusted copy number > 2 but < 10) and homozygous deletion had highly significant (Wilcoxon test, two-sided, P = 9.62 x 10-16 and P = 7.64 x 10-11, respectively) but less dramatic effects on expression with a lower penetrance (Fig. 2c). This lack of penetrance was associated with low cellularity as calculated by ASCAT (Wilcoxon test, two-sided, overexpression cut off = 2.5x normalised expression, P = 0.011) in non-extrachromosomal-like amplified cases but also likely reflects that specific genetic rearrangements, not just gene-dosage, can modulate expression. We also detected several cases of overexpression or complete expression loss without associated copy number changes, reflecting non-genetic mechanisms for driver dysregulation. One case overexpressed ERBB2 at 28-fold median expression but had entirely diploid copy number in and surrounding ERBB2, and a second case lost SMAD4 expression (0.008-fold median expression) despite possessing five copies of SMAD4.
Landscape of driver events in EAC
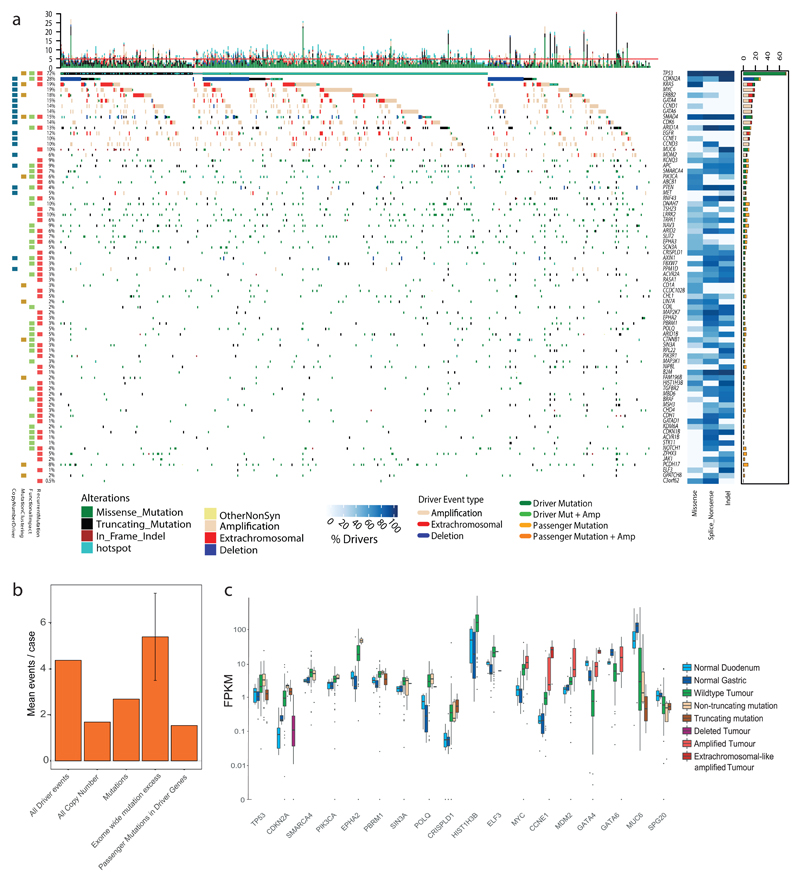
The overall landscape of driver gene mutations and copy number alterations per case is depicted in Figure 3a. These comprise both oncogenes and tumor suppressor genes activated or repressed via different mechanisms. Passenger mutations occur by chance in most driver genes. To quantify this, we used the observed:expected mutation ratios (calculated by dNdScv) to estimate the percentage of driver mutations in each gene and in different mutation classes. For many drivers, only specific mutation classes appear to be under selection. Many tumor suppressor genes (ARID2, RNF43, ARID1B for example) are only under selection for truncating mutations, i.e. splice site, nonsense and frameshift Indel mutations, but not missense mutations, which are passengers. However, oncogenes, like ERBB2, only contain missense drivers that form clusters to activate gene function in a specific manner. Where a mutation class is <100% driver mutations, mutational clustering can help us define the driver vs. passenger status of a mutation (Supplementary Fig. 6). Mutational hotspots in EAC or other cancer types32 (Supplementary Table 7 and Supplementary Data) are indicated in Figure 3a. Novel EAC drivers of particular interest include B2M, a core component of the MHC class I complex and a marker of acquired resistance to immunotherapy33, MUC6, a secreted glycoprotein involved in gastric acid resistance, and ABCB1, a channel pump protein associated with multiple instances of drug resistance34. We note that several of these drivers have been previously associated with gastric and colorectal cancer (Supplementary Table 8)13,35.
Figure 3. The driver gene landscape of EAC.
a, Driver mutations or CNVs are shown for each patient of 551 EACs. Amplification is defined as >2 copy number adjusted ploidy (2x ploidy of that case) and extrachromosomal amplification as >10 copy number adjusted ploidy (10x ploidy for that case). Driver associated features for each driver gene are displayed to the left. On the right, the percentages of different mutation and copy number changes are displayed, differentiating between driver and passenger mutations using dNdScv, and the % of predicted drivers by mutation type is shown. Above the plot are the number of driver mutations per sample with an indication of the mean (red line = 5). b, Mean driver events per case in 551 EACsand comparison to exome-wide excess of mutations generated by dNdScv. c, Expression changes in EAC driver genes in comparison to normal intestinal tissues in RNA matched samples (n=116). Only genes with expression changes of note are shown.
The identification of driver events provides rich information about the molecular history of each EAC tumor. We detect a median of five events in driver genes per tumor (IQR = 3-7, mean = 5.6), and only a very small fraction of cases has no such events detected (6 cases, 1%). When we remove the predicted percentage of passenger mutations using observed:expected mutation ratios calculated by dNdScv, one of the driver gene detection methods used, we find a mean of 4.4 true driver events per case. These derive more commonly from mutations than copy number events (Fig. 3b and Supplementary Table 9). Using hierarchal clustering of drivers, we noted that TP53 mutant cases had significantly more copy number drivers (Wilcoxon test, two-sided, P = 0.0032, Supplementary Figs. 7 and 8). dNdScv also analyses the genome-wide excess of non-synonymous mutations based on dN/dS ratios to assess the mean number of exonic driver mutations per case. This is calculated at 5.4 (95% CIs: 3.5-7.3) in comparison to a mean excess of 2.7 driver mutations in specific EAC driver genes, suggesting further low frequency driver genes are yet to be discovered in EAC.
To better understand the functional impact of driver mutations, we analyzed expression of driver genes with different mutation types and compared their expression to normal tissue RNA (Fig. 3c and Supplementary Fig. 10). Since surrounding squamous epithelium is a fundamentally different tissue from which EAC does not directly arise, we have used duodenum and gastric cardia samples as gastrointestinal phenotype controls, likely to be similar to the, as yet unconfirmed, tissue of origin in EAC. A large number of driver genes have upregulated expression in comparison to normal controls; for example, TP53 has upregulated RNA expression in wild-type tumor tissue and in cases with non-truncating mutations but RNA expression is lost upon gene truncation. In depth analysis of different TP53 mutation types reveals significant heterogeneity within non-truncating mutations (Supplementary Fig. 9). Normal tissue expression of CDKN2A suggests that CDKN2A is generally activated in EAC, likely due to genotoxic or other cancer-associated cellular stresses36, and returns to physiologically normal levels when deleted. Heterogeneous expression in wild-type CDKN2A cases suggests a different mechanism of inhibition, perhaps methylation, in some cases. Overexpression of some oncogenes occurs without genomic aberrations, such as MYC, which is overexpressed in MYC-wild-type EACs relative to normal tissues (Fig. 3c). A smaller number of driver genes are downregulated in EACs without genomic aberrations. 3/4 of these genes (GATA4, GATA6 and MUC6) are involved in the differentiated phenotype of gastrointestinal tissues and may be lost with tumor de-differentiation.
Dysregulation of specific pathways and processes in EAC
It is known that selection preferentially dysregulates certain functionally related groups of genes and biological pathways in cancer37. This phenomenon is highly evident in EAC, as shown in Figure 4, which depicts the functional relationships between EAC drivers (Supplementary Note). While TP53 is the dominant driver in EAC, 28% of cases remain TP53 wild-type. MDM2 is a E3 ubiquitin ligase that targets TP53 for degradation. Its selective amplification and overexpression is mutually exclusive with TP53 mutation, suggesting it can functionally substitute the effect of TP53 mutation via its degradation. Similar mutually exclusive relationships are observed between KRAS and ERBB2, GATA4 and GATA6, and cyclin genes (CCNE1, CCND1 and CCND3). Activation of the Wnt pathway occurs in 19% of cases either by mutation of phospho-residues at the N terminus of β-catenin, which prevent degradation, or loss of Wnt destruction complex components like APC. Many different chromatin modifying genes, often belonging to the SWI/SNF complex, are also selectively mutated (28% of cases). In contrast to other pathways, SWI/SNF genes are co-mutated significantly more often than we would expect by chance (Fisher’s exact test, two-sided, q < 0.05 for each gene; see Methods), suggesting these mutations are synergistic. We also assessed mutual exclusivity and co-occurrence in genes in different pathways and between pathways themselves (Fig. 4b). Of particular note are co-occurring relationships between TP53 and MYC, GATA6 and SMAD4, and Wnt and immune pathways, as well as mutually exclusive relationships between ARID1A and MYC, gastrointestinal (GI) differentiation and RTK pathways, and SWI-SNF and DNA damage response pathways. We were able to confirm some of these relationships in independent cohorts in different cancer types (Supplementary Table 10), suggesting some of these may be pan-cancer phenomenon. Wnt dysregulation was associated with hyper-mutated cases (> 500 exonic SNVs or Indels, Fisher’s exact test, two-sided, P = 2.98 x 10-5, OR = 9.3), as was mutation in immune pathway genes (B2M and JAK1, > 500 exonic SNVs or Indels, Fisher’s exact test, two-sided, P = 6.27 x 10-6, OR = 35.7).
Figure 4. Biological pathways undergoing selective dysregulation in EAC.
a, Biological pathways dysregulated by driver gene mutation and/or CNVs in 551 cases. Wild-type cases for a pathway are not shown. Inter and intra-pathway interactions are described, and mutual exclusivities and/or associations between genes in a pathway are annotated. GATA4 and GATA6 amplifications have a mutually exclusive relationship, although this does not reach statistical significance (Fisher’s exact test, two-sided, P = 0.07, OR = 0.52). b, Pairwise assessment of mutual exclusivity and association in EAC driver genes and pathways. Two sided Fisher’s exact test were used and hyper-mutated (>500 exonic mutations) cases were removed to avoid bias towards co-occurrence, hence n = 510. RTK; Receptor Tyrosine Kinase pathway.
EAC driver events correlate with clinical phenotype
Events undergoing selection during cancer evolution influence tumor biology and thus impact tumor aggressiveness, response to treatment, and patient prognosis, as well as other clinical parameters.
Univariate Cox regression was performed for events in each driver gene with driver events occurring in greater than 5% of EACs after passenger removal to detect prognostic biomarkers (Fig. 5a). Events in two genes conferred significantly poorer prognosis after multiple hypothesis correction: GATA4 amplification (HR = 0.54, 95% CI = 0.38–0.78, P = 0.0008) and SMAD4 mutation or homozygous deletion (HR = 0.60, 95% CI = 0.42–0.84, P = 0.003), which were present in 31% of EACs (Fig. 5b). Both genes remained significant in multivariate Cox regression, including pathological TNM staging, resection margin, curative vs. palliative treatment intent, and differentiation status (GATA4, HR adjusted = 0.47, 95% CIs adjusted = 0.29–0.76, P = 0.002; SMAD4, HR adjusted = 0.61, 95% CI adjusted = 0.40–0.94, P = 0.026) (Fig. 5b, Supplementary Fig. 11). We validated the poor prognostic impact of SMAD4 events in an independent TCGA gastroesophageal cohort (HR = 0.58, 95% CI = 0.37–0.90, P = 0.014) (Fig. 5c), and we also found GATA4 amplifications were prognostic in a cohort of TCGA pancreatic cancers (HR = 0.38, 95% CI = 0.18–0.80, P = 0.011) (Fig. 5d), the only available cohort containing a feasible number of GATA4 amplifications. The prognostic impact of GATA4 has been suggested in previously published independent EAC cohort16, although it did not reach statistical significance after FDR correction, and SMAD4 expression loss has been previously linked to poor prognosis in EAC38. We also noted stark survival differences between cases with SMAD4 events and cases in which TGFβ receptors were mutated (Fig. 5e, HR = 5.6, 95% CI = 1.7–18.2, P = 0.005), in keeping with the biology of the TGFβ pathway, where non-SMAD TGFβ signalling is known to be oncogenic39.
Figure 5. Clinical significance of driver events in 379 clinically annotated EACs.
a, Hazard ratios and 95% confidence intervals for Cox regression analysis across all driver genes with at least a 5% frequency of driver alterations. *q < 0.05 after BH adjustment. b, Kaplan-Meier curves for EACs with different status of significant prognostic indicators (GATA4 and SMAD4). c, Kaplan-Meier curves for different alterations in the TGF-β pathway. d, Kaplan-Meier curves showing verification GATA4 prognostic value in GI cancers using a pancreatic TCGA cohort. e, Kaplan-Meier curves showing verification SMAD4 prognostic value in gastroesophageal cancers using a gastroesophageal TCGA cohort. f, Differentiation bias in tumors containing events in Wnt pathway driver genes. g, Relative frequency of KRAS mutations and TP53 mutations driver gene events in females vs. males (Fisher’s exact test, two sided).
In additional to survival analyses, we also assessed driver gene events for correlation with various other clinical factors, including differentiation status, sex, age and treatment response. We found Wnt pathway mutations had a strong association with well differentiated tumours (P = 0.001, OR = 2.9, Fisher’s test, two-sided, see Methods; Fig. 5f). Female cases (n = 81) were enriched for KRAS mutation (P = 0.001, Fisher’s exact test, two-sided) and TP53 wild-type status (P = 0.006, Fisher’s exact test, two-sided) (Fig. 5g). This is of particular interest given the male predominance of EAC3.
Targeted therapeutics using EAC driver events
To investigate whether driver events in particular genes and/or pathways might sensitize EAC cells to certain targeted therapeutic agents, we used the Cancer Biomarkers database40. We calculated the percentage of our cases that contain EAC-driver biomarkers of response to each drug class in the database (Fig. 6a, and full data in Supplementary Table 11). Aside from TP53, which has been problematic to target clinically so far, we found a number of drugs with predicted sensitivity in >10% of EACs, including EZH2 inhibitors for some SWI/SNF mutant cancers (23%, and 28% including all SWI/SNF EAC drivers), and BET inhibitors, which target KRAS activated and MYC amplified cases (25%). However, by far the most significantly effective drug was predicted to be CDK4/6 inhibitors, where >50% of cases harbored sensitivity causing events in the receptor tyrosine kinase (RTK) and core cell cycle pathways (e.g. in CCND1, CCND3 and KRAS).
Figure 6. CDK4/6 inhibitors in EAC.
a, Drug classes for which sensitivity is indicated by EAC driver genes with data from the Cancer Biomarkers database36. b, Area under the curve (AUC) of sensitivity is shown in a panel of 13 EAC and Barrett’s esophagus high grade dysplasia cell lines with associated WGS and their corresponding driver events, based on primary tumor analysis. AUC is also shown for two control lines: T47D, an ER-positive breast cancer line (positive control), and MDA-MB-468, an Rb negative breast cancer (negative control). *CCNE1 is a known marker of resistance to CDK4/6 inhibitors due to its regulation of Rb downstream of CDK4/6, hence bypassing the need for CDK4/6 activity (see Fig. 4). c, Response of organoid cultures to three FDA approved CDK4/6 inhibitors and corresponding driver events. RTK; Receptor tyrosine kinase pathway, BC; Breast Cancer.
To verify that these driver events would also sensitize EAC tumors to such inhibitors, we used a panel of 13 EAC or Barrett’s high grade dysplasia cell lines that have undergone whole genome sequencing41 and assessed them for presence of EAC driver events (Fig. 6b). The mutational landscape of these lines was broadly representative of EAC tumors. We found that the presence of cell cycle and or RTK activating driver events was highly correlated with response to two FDA approved CDK4/6 inhibitors, Ribociclib and Palbociclib, and several cell lines were sensitive below maximum tolerated blood concentrations in humans (Fig. 6b, Supplementary Table 12, and Supplementary Fig. 12)42. Such EAC cell lines had comparable sensitivity to T47D, which is derived from an ER-positive breast cancer, where CDK4/6 inhibitors have been FDA approved. We noted three cell lines that were highly resistant, with little drug effect even at 4,000 nM concentrations, similar to a known Rb mutant resistant line breast cancer cell line (MDA-MB-468). Two of these three cell lines harbor amplification of CCNE1, which is known to drive resistance to CDK4/6 inhibitors by bypassing CDK4/6 and causing Rb phosphorylation via CDK2 activation43. To verify these effects in a more representative model of EAC, we treated three whole genome sequenced EAC organoid cultures44 with Palbociclib and Ribociclib as well as a more recently approved CDK4/6 inhibitor, Abemaciclib. As was observed in cell lines, cell cycle and RTK driver events were present only in the more sensitive organoids and CCNE1 activation in the most resistant (Fig. 6c).
Discussion
We present here a detailed catalog of coding and non-coding genomic events that have been selected for during the evolution of esophageal adenocarcinoma. These events have been characterized in terms of their relative impact, related functions, mutual exclusivity and co-occurrence and expression in comparison to normal tissues. We have used this set of biologically important gene alterations to identify prognostic biomarkers and actionable genomic events for personalized medicine.
While clinical annotation and matched RNA data is a strength of this study, in some cases we may have been unable to assess selected variants expression changes that were detected in the full 551 cohort due to lack of representation RNA matched sub-cohort. Despite rigorous analyses to detect selected events, assessment of the global excess of mutations by dNdScv suggests that we are unable to detect all mutations selected in EAC, similar to many other cancer types21. All driver gene detection methods that we have used rely on driver mutation re-occurrence in a genomic region to some degree. Many of these undetected driver mutations are hence likely to be spread across a large number of genes, whereby each is mutated at very low frequency across EAC patients. This tendency for low frequency EAC drivers may be responsible for the low yield of MutSigCV in previous cohorts and may suggests that C-type cancers such as EAC are not less ‘mutation-driven’ than M-type cancers but rather that their mutational drivers are spread across a larger number of genes5. Copy number driver gene identification is even more challenging due to the large size and lower frequency of these events, and hence it is also possible that there are significantly more EAC copy number drivers yet to discovered, possibly already identified as candidates here.
While a number of previous reports have attempted to detect EAC drivers, they have had a limited yield per case. The first such study19 used methods that, despite being well regarded at the time, were subsequently discredited8. Since then, a number of reports, including our own, on medium and large cohort sizes using MutSigCV9,10,17 were only able to detect a small number of mutational driver genes (7, 5 and 15 in each study). By using both a large cohort and more comprehensive methodologies, we have significantly increased this figure to 66 mutational driver genes (excluding copy number drivers). Detection of driver CNAs has previously relied on GISTIC to detect recurrently copy number aberrant regions9,14–17, but no analyses have been performed to determine which genes in these large regions are true drivers. Many of the genes annotated by such papers are unlikely to be copy number drivers due to their lack of expression modulation with CNAs (e.g. YEATS4 and MCL1), the role of recurrent heterozygous losses to drive LOH in some mutational drivers (ARID1A and CDH11) or their association with fragile sites (PDE4D, WWOX, FHIT). Conversely, we have been able to identify novel EAC copy number drivers (e.g. CCND3, AXIN1, PPM1D and APC).
We have noted a three-way association between hyper-mutation, Wnt activation and loss of immune signalling genes such as B2M. MSI-driven hyper-mutation has been previously associated with higher immune activity45,46. However, Wnt dysregulation and mutation of immune pathway genes such as B2M33 have been previously linked to immune escape47, suggesting this may be an acquired mechanism to prevent immune surveillance caused by hyper-mutation.
Functional characterization of many of the driver genes described is needed to understand why they are advantageous to EAC tumors and how they modify EAC biology. Biological pathways and processes that are selectively dysregulated deserve particular attention in this regard, as do the gene pairs or groups with mutually exclusive or co-occurring relationships such as MYC and TP53 or SWI/SNF factors, suggestive of particular functional relationships. Prospective clinical work to verify and implement SMAD4 and GATA4 biomarkers in this study would be worthwhile. While EAC is a poor prognosis cancer type, significant heterogeneity of survival outcome makes triaging patients in treatment groups an important part of clinic practice, which could be improved using better prognostication. A number of targeted therapeutics may provide clinic benefit to EAC cases based on their individual genomic profile. In particular, CDK4/6 inhibitors deserve considerable attention as an option for EAC treatment as they are, by a significant margin, the treatment to which the most EACs harbor sensitivity-causing driver events, excluding TP53 as an unlikely therapeutic biomarker. Previous work has noted activity of the CDK4/6 inhibitor Palbociclib in a small number of EAC cell lines48, but biomarkers were not investigated. The extensive in vitro validation of identified biomarkers for CDK4/6 inhibitors in EAC across 16 cell lines and organoids is persuasive of possible clinical benefit using a targeted approach.
In summary, this work provides a detailed compendium of mutations and copy number alterations undergoing selection in EAC, which have clinically relevant impact on tumor behaviour. This comprehensive study provides us with useful insights into the nature of EAC tumors and should pave the way for evidence based clinical trials in this poor prognosis disease.
Methods
Cohort, sequencing and calling of genomic events
379 cases (69%) of our EAC cohort were derived from the esophageal adenocarcinoma WGS ICGC study, for which samples are collected through the UK wide OCCAMS (Oesophageal Cancer Classification and Molecular Stratification) consortium. The procedures for obtaining the samples, quality control processes, extractions and whole genome sequencing are as previously described17. Strict pathology consensus review was observed for these samples with a 70% cellularity requirement before inclusion. Comprehensive clinical information was available for the ICGC-OCCAMS cases (Supplementary Table 13). In addition, previously published samples were included in the analysis from Dulak et al.19 (149 WES; 27%) and Nones et al.20 (22 WGS samples; 4%) to total 551 genome characterized EACs. RNA-seq data was available from our ICGC WGS samples (116/379). BAM files for all samples (include those from Dulak et al.19 and Nones et al.20) were run through our alignment (BWA-MEM), mutation (Strelka), copy number (ASCAT) and structural variant (Manta) calling pipelines, as previously described17. Our methods were benchmarked against various other available methods and have among the best sensitivity and specificity for variant calling (ICGC benchmarking exercise49,50). Cell lines were whole genome sequenced at 30X coverage with 150bp paired end reads on an Illumina Hiseq4000. Copy number calling was performed by Freec as previously described41. Mutations were called by GATK as previously described41, filtered for germline variants in the 1000 genomes project and any known oncogenic hotspots32 were recovered. Amplifications were defined as genes with 2x the median copy number of the host chromosome or greater.
Total RNA was extracted using All Prep DNA/RNA kit from Qiagen, and the quality was checked on Agilent 2100 Bioanalyzer using RNA 6000 nano kit (Agilent). Qubit High sensitivity RNA assay kit from Thermo Fisher was used for quantification. Libraries were prepared from 250 ng RNA, using TruSeq Stranded Total RNA Library Prep Gold (Ribo-zero) kit, and ribosomal RNA (nuclear, cytoplasmic and mitochondrial rRNA) was depleted, whereby biotinylated probes selectively bind to ribosomal RNA molecules forming probe-rRNA hybrids. These hybrids were pulled down using magnetic beads and rRNA depleted total RNA was reverse transcribed. The libraries were prepared according to Illumina protocol51. Paired end 75-bp sequencing on HiSeq4000 generated the paired end reads. For normal expression controls, we chose gastric cardia tissue, from which some hypothesize Barrett’s esophagus may arise, and duodenum which contains intestinal histology, including goblet cells, which mimics that of Barrett’s esophagus. We did not use Barrett’s esophagus tissue itself as a normal control given the heterogeneous and plentiful phenotypic and genomic changes that it undergoes early in its pathogenesis.
Analyzing EAC mutations for selection
To detect positively selected mutations in our EAC cohort, a multi-tool approach across various selection related ‘features’ (recurrence, functional impact, clustering) was implemented in order to provide a comprehensive analysis. This is broadly similar to several previous approaches7,11. dNdScv21, MutSigCV8, e-Driver25, ActivedriverWGS22 and e-Driver3D26 were run using the default parameters. To run OncodriverFM23, Polyphen52 and SIFT53 were used to score the functional impact of each missense non-synonymous mutation (from 0 non-impactful to 1 highly impactful); synonymous mutations were given a score of 0 impact, and truncating mutations (nonsense and frameshift mutations) were given a score of 1. Any gene with less than 7 mutations, unlikely to contain detectable drivers using this method, was not considered to decrease the false discovery rate (FDR). OncodriveClust was run using a minimum cluster distance of 3, minimum number of mutations for a gene to be considered of 7 and with a stringent probability cut off to find cluster seeds of P = 1 x 10-13 to prevent infiltration of large numbers of, likely, false positive genes. For all tool outputs, we undertook quality control including Q-Q plots to ensure no tool produces inflated q-values and each tool produced at least 30% known cancer genes. Two tools were removed from the analysis due to failure for both of these parameters at quality control in our hands (Activedriver54 and Hotspot32). For three of the QC-approved tools (dNdScv, OncodriveFM, MutSigCV) where this was possible, we also undertook an additional FDR reducing analysis by re-calculating q values based on analysis of known cancer genes only21,27,28 as has been previously implemented21,55. Significance cut offs were set at q < 0.1 for coding genes. Tool outputs were then put through various filters to remove any further possible false positive genes. Specifically, genes where <50% of EAC cases had no expression (TPM<0.1) in our matched RNA-seq cohort were removed and, using dNdScv, genes with no or only a small mutation excess (observed: expected ratio > 1.5:1) of any single mutation type were also removed. We also removed mitochondrial genes two (MT-MD2, MT-MD4) that were highly enriched for truncating mutations and were frequently called in OncodriveFM as well as other tools. This is may be due to the different mutational dynamics caused by ROS from the mitochondrial electron transport chain and the high number of mitochondrial genomes per cell, which enables significantly more heterogeneity. These factors prevent the tools used from calculating an accurate null model for these genes, but they may be worthy of functional investigation. ActiveDriverWGS calculates an expected background mutation rate based on mutation rates of local, adjacent sequence for each tested element while correcting for the differential mutation rates within each trinucleotide context. It thus tests observed mutation rates against this predicted background for each element. ActiveDriverWGS also detects elements with mutations enriched in binding site regions (high impact). For non-coding elements called by ActivedriverWGS, filtering for expression or dN/dS was not possible, and despite recent benchmarking29, such methods are not so well established. Hence we took a more cautious approach with general significance cut offs of q < 0.001 and q < 0.1 for previously identified elements in other cancer types11. q values were not recalculated for previously identified elements alone like with coding genes, but the q < 0.1 cut off was calculated based on P values for all assessed elements. To calculate exome-wide mutational excess, hyper-mutated cases (>500 exonic mutations) were removed and the global non-synonymous dN/dS ratios were applied to all dNdScv annotated mutations excluding “synonymous” and “no SNV” annotations as described in Martincorena et al.21.
Detecting selection in CNVs
ASCAT raw copy number values were used to detected frequently deleted or amplified regions of the genome using GISTIC2.014. To determine which genes in these regions confer a selective advantage, CNVs from each gene within GISTIC identified loci were correlated with TPM from matched RNA-seq in a sub-cohort of 116 samples and with mutations across all 551 samples. To call copy number in genes that spanned multiple copy number segments in ASCAT, we considered the total number of full copies of the gene (i.e. the lowest total copy number). Occasionally ASCAT is unable to confidently call the copy number in highly aberrant genomic regions. We found that the expression of genes in such regions matched well what we would expect given the surrounding copy number, and hence we used the mean of the two adjacent copy number fragments to call copy number in the gene in question. We found amplification peak regions identified by GISTIC2.0 varied significantly in precise location both in analysis of different sub-cohorts and when comparing to published GISTIC data from EACs9,15,16. A peak would often sit next to but not overlapping a well-characterized oncogene or tumor suppressor. To account for this, we widened the amplification peak sizes upstream and downstream by twice the size of each peak to ensure we captured all possible drivers. Our expression analysis allows us to then remove false positives from this wider region, and called drivers were still highly enriched for genes closer to the centre of GISTIC peak regions.
To detect genes in which amplification correlated with increased expression, we compared expression of samples with a high copy number for that gene (above 10th percentile CN/ploidy) with those that have a normal copy number (median +/- 1) using the Wilcoxon rank-sum test and using the specific alternative hypothesis that high copy number would lead to increased expression. q-values were then generated based on the Benjamini and Hochberg method, not considering genes without significant expression in amplified samples (at least 75% amplified samples with TPM > 0.1) and considering q < 0.001 as significant. We also included an additional known driver gene only FDR reduction analysis as previously described for mutational drivers, with q < 0.1 considered as significant given the additional evidence for these genes in other cancer types. We also included MYC despite its P = 0.11 for expression correlation. This is due to frequent non-amplification associated overexpression of MYC when compared to normal controls, and otherwise MYC is well evidenced by a very close proximity to the peak centre (top 4 genes) and its high rate of amplification (19%). We took the same approach to detect genes in which homozygous deletion correlated with expression loss, comparing cases with copy number = 0 to all others. Large expression modulation was a highly specific marker for known copy number driver genes and was not a widespread feature in most recurrently copy number variant genes. However, while expression modulation is a requirement for selection of CNV only drivers, it is not sufficient evidence alone, and hence we grouped such genes into those which have been characterized as drivers previously in other cancer types (high confidence EAC copy number drivers) and other genes (candidate EAC copy number drivers), which await functional validation. We used fragile site regions detected in Wala et al.56. We also defined regions that may be recurrently heterozygous deleted, without any significant expression modulations, to allow LOH of tumor suppressor gene mutations. To do this, we analyzed genes with at least 5 mutations for association between LOH (ASCAT minor allele = 0) and mutation using Fisher’s exact test and generated q values using the Benjamini and Hochberg method. The analysis was repeated on known cancer genes only for reduced FDR and q < 0.1 considered significant for both analyses. For those high confidence drivers, we chose to define amplification as total copy number/ploidy (referred to as ploidy adjusted copy number) because this produces superior correlation with expression. We chose a cut off for amplification at ploidy adjusted copy number = 2 as has been previously used, and causes a highly significant increase in expression in our copy number-driver genes when amplified.
Pathways and relative distributions of genomic events
The relative distribution of driver events in each pathway was analysed using a Fisher’s exact test in the case of pair-wise comparisons including wild-type cases. In the case of multi-gene comparisons such as cyclins, we calculate the P value and odds ratio for gene in the group using a two-sided Fisher’s exact test, corrected by Benjamini and Hochberg, and combine resulting q values using the Fisher method; genes without odds ratios > 2 for co-occurrence and < 0.5 for mutual exclusivity were removed. For this analysis, we also remove highly mutated cases (>500 exonic mutations, 41/551) as they bias distribution of genes towards co-occurrence. To ensure that a non-random distribution of mutations across samples was still not affecting the strong co-occurrence of SWI/SNF genes (all genes q < 0.05 before combining q values), we repeated the analysis randomly iterating 30,000 times over other driver gene eight combinations (excluding SWI/SNF genes) and found only 0.01% (4/30,000) of random combinations had all genes q < 0.05 as found in SWI/SNF genes. We then performed this analyses across all pairs of driver genes using two sided Fisher’s exact tests and Benjamini and Hochberg multiple hypothesis correction (q values < 0.1 are shown in Fig. 4b). We validated these relationships in independent TGCA cohorts of other GI cancers where we could find cohorts with reasonable numbers of the genomic events in question (not possible for GATA4/GATA6, for instance) using the cBioportal web interface tool57.
Correlating genomics with the clinical phenotype
To find genomic markers for prognosis, we undertook univariate Cox regression for those driver genes present in >5% of cases (n = 16) along with Benjamini and Hochberg false discovery correction. We considered only these genes to reduce our false discovery rate and because other genes were unlikely to impact on clinical practice given their low frequency in EAC. We validated SMAD4 in the TCGA gastroesophageal cohort, which had a comparable frequency of these events, but notably is composed mainly of gastric cancers, and GATA4 in the TCGA pancreatic cohort using the cBioportal web interface tool. We also validated these markers as independent predictors of survival both in respect of each other and stage using a multivariate Cox regression in our 379 clinical annotated ICGC cohort. When assessing for genomic correlates with differentiation phenotypes, we found only very few cases with well differentiated phenotypes (<5% cases), and hence for statistical analyses, we collapse these cases with moderate differentiation to allow a binary Fisher’s exact test to compare poorly differentiated with well-moderate differentiated phenotypes.
Therapeutics
The cancer biomarker database was filtered for drugs linked to biomarkers found in EAC drivers, and Supplementary Table 8 was constructed using the cohort frequencies of EAC biomarkers. Ten EAC cell lines (SKGT4, OACP4C, OACM5.1, ESO26, ESO51, OE33, MFD, OE19, Flo-1 and JHesoAD) and three Barrett’s esophagus high grade dysplasia cell lines (CP-B, CP-C and CP-D) with WGS data41 were used in proliferation assays to determine drug sensitivity to CDK4/6 inhibitors, Palbociclib (Biovision) and Ribociclib (Selleckchem). Cell lines were grown in their normal growth media. Proliferation was measured using the Incucyte live cell analysis system (Incucyte ZOOM Essen biosciences). Each cell line was plated at a starting confluency of 10% and growth rate measured across 4-7 days depending on basal proliferation rate (until 90% confluent). For each cell-line drug combination, concentrations of 16, 64, 250, 1,000 and 4,000 nM were used each in 0.3% DMSO and compared to 0.3% DMSO only. Each condition was performed in at least triplicate (technical replicates) and 12/12 randomly chosen cell line; drug combinations were successfully replicated with biological replicates (independent experiments). The time period of treatment to growth cessation in the control (0.3% DMSO) condition was used to calculate GI50 and AUC. Accurate GI50s could not be calculated in cases where a cell line had >50% proliferation inhibition even with the highest drug concentration, and hence AUC was used to compare cell line sensitivity. T47D had a highly similar GI50 for Palbociclib to that previously calculated in other studies (112 nM vs. 127 nM)58. Primary organoid cultures were derived from EAC cases included in the OCCAMS/ICGC sequencing study. Detailed organoid culture and derivation method have been previously described44. Regarding the drug treatment, the seeding density for each organoid line was optimized to ensure cell growth in the logarithmic growth phase. Cells were seeded in complete medium for 24 hours then treated with compounds at 5-point 4-fold serial dilutions for 6 days or 12 days. Cell viability was assessed using CellTiter-Glo (Promega) after drug incubation.
Supplementary Material
Editorial summary.
Genomic analysis of 551 esophageal adenocarcinomas identifies new driver mutations and biomarkers associated with poor prognosis. Over 50% of esophageal adenocarcinomas contain sensitizing events for CDK4/6 inhibitors, providing an evidence base for targeted therapeutics.
Acknowledgements
We thank A. J. Bass and N. Waddell for providing data in Dulak et al.19 and Nones et al.20, respectively, also included in our previous publication18. Inclusion of this data allowed augmentation of our ICGC cohort and a greater sensitivity for the detection of EAC driver variants.
OCCAMS was funded by a programme grant from Cancer Research UK (RG66287), and the Fitzgerald laboratory is funded by a Core Programme Grant from the Medical Research Council. We thank the Human Research Tissue Bank, which is supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre, from Addenbrooke’s Hospital. Additional infrastructure support was provided from the CRUK funded Experimental Cancer Medicine Centre.
Footnotes
Data availability. The WGS and RNA expression data can be found at the European Genome-phenome Archive (EGA) under accessions EGAD00001004417 and EGAD00001004423, respectively.
Code availability. Code associated with the analysis is available upon request.
Ethics. The study was registered (UKCRNID 8880), approved by the Institutional Ethics Committees (REC 07/H0305/52 and 10/H0305/1), and all subjects gave individual informed consent.
Reporting summary. Additional information is included in the Life Sciences Reporting Summary, which details exact software and biological materials used and efforts made to ensure reproducibility of results.
Author contributions
RCF and AMF conceived the overall study. AMF and SJ analyzed the genomic data and performed statistical analyses. RCF, AMF and XL designed the experiments. AMF, XL and JM performed the experiments. GC contributed to the structural variant analysis and data visualization. SK helped compile the clinical data and aided statistical analyses. JP and SA produced and QC’ed the RNA-seq data. EO aided the whole genome sequencing of EAC cell lines. SM and NG coordinated the clinical centres and were responsible for sample collections. ME benchmarked our mutation calling pipelines. MO led the pathological sample QC for sequencing. LB and GD constructed and managed the sequencing alignment and variant calling pipelines. RCF and ST supervised the research. RCF and ST obtained funding. AMF and RCF wrote the manuscript. All authors approved the manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015 Mar 1;136(5):E359–386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Coleman HG, Xie SH, Lagergren J. The Epidemiology of Esophageal Adenocarcinoma. Gastroenterology. 2018 Jan;154(2):390–405. doi: 10.1053/j.gastro.2017.07.046. [DOI] [PubMed] [Google Scholar]
- 3.Smyth EC, Lagergren J, Fitzgerald RC, et al. Oesophageal cancer. Nat Rev Dis Primers. 2017 Jul 27;3:17048. doi: 10.1038/nrdp.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell PJ, Getz G, Stuart JM, Korbel JO, Stein LD. Pan-cancer analysis of whole genomes. bioRxiv. 2017 [Google Scholar]
- 5.Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013 Oct;45(10):1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Secrier M, Li X, de Silva N, et al. Mutational signatures in esophageal adenocarcinoma define etiologically distinct subgroups with therapeutic relevance. Nat Genet. 2016 Oct;48(10):1131–1141. doi: 10.1038/ng.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamborero D, Gonzalez-Perez A, Perez-Llamas C, et al. Comprehensive identification of mutational cancer driver genes across 12 tumor types. Sci Rep. 2013 Oct 2;3:2650. doi: 10.1038/srep02650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence MS, Stojanov P, Polak P, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013 Jul 11;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Integrated genomic characterization of oesophageal carcinoma. Nature. 2017 Jan 12;541(7636):169–175. doi: 10.1038/nature20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin DC, Dinh HQ, Xie JJ, et al. Identification of distinct mutational patterns and new driver genes in oesophageal squamous cell carcinomas and adenocarcinomas. Gut. 2017 Aug 31; doi: 10.1136/gutjnl-2017-314607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rheinbay E, Nielsen MM, Abascal F, et al. Discovery and characterization of coding and non-coding driver mutations in more than 2,500 whole cancer genomes. bioRxiv. 2017 [Google Scholar]
- 12.Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014 Mar 20;507(7492):315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014 Sep 11;513(7517):202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dulak AM, Schumacher SE, van Lieshout J, et al. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res. 2012 Sep 1;72(17):4383–4393. doi: 10.1158/0008-5472.CAN-11-3893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frankel A, Armour N, Nancarrow D, et al. Genome-wide analysis of esophageal adenocarcinoma yields specific copy number aberrations that correlate with prognosis. Genes Chromosomes Cancer. 2014 Apr;53(4):324–338. doi: 10.1002/gcc.22143. [DOI] [PubMed] [Google Scholar]
- 17.Secrier M, Fitzgerald RC. Signatures of Mutational Processes and Associated Risk Factors in Esophageal Squamous Cell Carcinoma: A Geographically Independent Stratification Strategy? Gastroenterology. 2016 May;150(5):1080–1083. doi: 10.1053/j.gastro.2016.03.017. [DOI] [PubMed] [Google Scholar]
- 18.Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013 Oct;45(10):1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dulak AM, Stojanov P, Peng S, et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat Genet. 2013 May;45(5):478–486. doi: 10.1038/ng.2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nones K, Waddell N, Wayte N, et al. Genomic catastrophes frequently arise in esophageal adenocarcinoma and drive tumorigenesis. Nat Commun. 2014 Oct 29;5 doi: 10.1038/ncomms6224. 5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martincorena I, Raine KM, Gerstung M, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017 Nov 16;171(5):1029–1041 e1021. doi: 10.1016/j.cell.2017.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wadi L, Uuskula-Reimand L, Isaev K, et al. Candidate cancer driver mutations in super-enhancers and long-range chromatin interaction networks. bioRxiv. 2017 doi: 10.1016/j.molcel.2019.12.027. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalez-Perez A, Lopez-Bigas N. Functional impact bias reveals cancer drivers. Nucleic Acids Res. 2012 Nov;40(21):e169. doi: 10.1093/nar/gks743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tamborero D, Gonzalez-Perez A, Lopez-Bigas N. OncodriveCLUST: exploiting the positional clustering of somatic mutations to identify cancer genes. Bioinformatics. 2013 Sep 15;29(18):2238–2244. doi: 10.1093/bioinformatics/btt395. [DOI] [PubMed] [Google Scholar]
- 25.Porta-Pardo E, Godzik A. e-Driver: a novel method to identify protein regions driving cancer. Bioinformatics. 2014 Nov 1;30(21):3109–3114. doi: 10.1093/bioinformatics/btu499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Porta-Pardo E, Hrabe T, Godzik A. Cancer3D: understanding cancer mutations through protein structures. Nucleic Acids Res. 2015 Jan;43(Database issue):D968–973. doi: 10.1093/nar/gku1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Futreal PA, Coin L, Marshall M, et al. A census of human cancer genes. Nat Rev Cancer. 2004 Mar;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013 Oct 17;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shuai S, Gallinger S, Stein LD. DriverPower: Combined burden and functional impact tests for cancer driver discovery. bioRxiv. 2017 doi: 10.1038/s41467-019-13929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor AM, Shih J, Ha G, et al. Genomic and Functional Approaches to Understanding Cancer Aneuploidy. Cancer cell. 2018 Apr 9;33(4):676–689 e673. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner KM, Deshpande V, Beyter D, et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature. 2017 Mar 2;543(7643):122–125. doi: 10.1038/nature21356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang MT, Asthana S, Gao SP, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016 Feb;34(2):155–163. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med. 2016 Sep 1;375(9):819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z, Shi T, Zhang L, et al. Mammalian drug efflux transporters of the ATP binding cassette (ABC) family in multidrug resistance: A review of the past decade. Cancer Lett. 2016 Jan 1;370(1):153–164. doi: 10.1016/j.canlet.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 35.Giannakis M, Mu XJ, Shukla SA, et al. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell reports. 2016 Oct 18;17(4):1206. doi: 10.1016/j.celrep.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pei XH, Xiong Y. Biochemical and cellular mechanisms of mammalian CDK inhibitors: a few unresolved issues. Oncogene. 2005 Apr 18;24(17):2787–2795. doi: 10.1038/sj.onc.1208611. [DOI] [PubMed] [Google Scholar]
- 37.Leiserson MD, Vandin F, Wu HT, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet. 2015 Feb;47(2):106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singhi AD, Foxwell TJ, Nason K, et al. Smad4 loss in esophageal adenocarcinoma is associated with an increased propensity for disease recurrence and poor survival. Am J Surg Pathol. 2015 Apr;39(4):487–495. doi: 10.1097/PAS.0000000000000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Levy L, Hill CS. Alterations in components of the TGF-beta superfamily signaling pathways in human cancer. Cytokine Growth Factor Rev. 2006 Feb-Apr;17(1–2):41–58. doi: 10.1016/j.cytogfr.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 40.Tamborero D, Rubio-Perez C, Deu-Pons J, et al. Cancer Genome Interpreter Annotates The Biological And Clinical Relevance Of Tumor Alterations. bioRxiv. 2017 doi: 10.1186/s13073-018-0531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Contino G, Eldridge MD, Secrier M, et al. Whole-genome sequencing of nine esophageal adenocarcinoma cell lines. F1000Res. 2016;5:1336. doi: 10.12688/f1000research.7033.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liston DR, Davis M. Clinically Relevant Concentrations of Anticancer Drugs: A Guide for Nonclinical Studies. Clin Cancer Res. 2017 Jul 15;23(14):3489–3498. doi: 10.1158/1078-0432.CCR-16-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herrera-Abreu MT, Palafox M, Asghar U, et al. Early Adaptation and Acquired Resistance to CDK4/6 Inhibition in Estrogen Receptor-Positive Breast Cancer. Cancer Res. 2016 Apr 15;76(8):2301–2313. doi: 10.1158/0008-5472.CAN-15-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, Francies HE, Secrier M, et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nature communications. 2018 Jul 30;9(1):2983. doi: 10.1038/s41467-018-05190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Llosa NJ, Cruise M, Tam A, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer discovery. 2015 Jan; 5(1):43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le DT, Uram JN, Wang H, et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. The New England journal of medicine. 2015 Jun 25;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grasso CS, Giannakis M, Wells DK, et al. Genetic Mechanisms of Immune Evasion in Colorectal Cancer. Cancer discovery. 2018 Jun;8(6):730–749. doi: 10.1158/2159-8290.CD-17-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ismail A, Bandla S, Reveiller M, et al. Early G(1) cyclin-dependent kinases as prognostic markers and potential therapeutic targets in esophageal adenocarcinoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011 Jul 1;17(13):4513–4522. doi: 10.1158/1078-0432.CCR-11-0244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ding J, McConechy MK, Horlings HM, et al. Systematic analysis of somatic mutations impacting gene expression in 12 tumour types. Nat Commun. 2015 Oct 5;6 doi: 10.1038/ncomms9554. 8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee AY, Ewing AD, Ellrott K, et al. Combining accurate tumor genome simulation with crowdsourcing to benchmark somatic structural variant detection. Genome Biol. 2018 Nov 6;19(1):188. doi: 10.1186/s13059-018-1539-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nagai K, Kohno K, Chiba M, et al. Differential expression profiles of sense and antisense transcripts between HCV-associated hepatocellular carcinoma and corresponding non-cancerous liver tissue. Int J Oncol. 2012 Jun;40(6):1813–1820. doi: 10.3892/ijo.2012.1382. [DOI] [PubMed] [Google Scholar]
- 52.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013 Jan; doi: 10.1002/0471142905.hg0720s76. Chapter 7:Unit7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ng PC, Henikoff S. Predicting the effects of amino acid substitutions on protein function. Annu Rev Genomics Hum Genet. 2006;7:61–80. doi: 10.1146/annurev.genom.7.080505.115630. [DOI] [PubMed] [Google Scholar]
- 54.Reimand J, Wagih O, Bader GD. The mutational landscape of phosphorylation signaling in cancer. Sci Rep. 2013 Oct 2;3 doi: 10.1038/srep02651. 2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Northcott PA, Buchhalter I, Morrissy AS, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017 Jul 19; 547(7663):311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wala JA, Shapira O, Li Y, et al. Selective and mechanistic sources of recurrent rearrangements across the cancer genome. bioRxiv. 2017 [Google Scholar]
- 57.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013 Apr 2;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Finn RS, Dering J, Conklin D, et al. PD 0332991, a selective cyclin D kinase 4/6 inhibitor, preferentially inhibits proliferation of luminal estrogen receptor-positive human breast cancer cell lines in vitro. Breast Cancer Res. 2009;11(5):R77. doi: 10.1186/bcr2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.