Abstract
Alcohol use disorder (AUD) is one of the most common preventable mental health disorders and can result in pathology within the CNS, including the cerebellum. Cerebellar alcohol exposure during adulthood has been associated with disruptions in proper cerebellar function. However, the mechanisms regulating ethanol-induced cerebellar neuropathology are not well understood. High-throughput next generation sequencing was performed to compare control versus ethanol-treated adult C57BL/6J mice in a chronic plus binge model of AUD. Mice were euthanized, cerebella were microdissected, and RNA was isolated and submitted for RNA-sequencing. Down-stream transcriptomic analyses revealed significant changes in gene expression and global biological pathways in control versus ethanol-treated mice that included pathogen-influenced signaling pathways and cellular immune response pathways. Microglial-associated genes showed a decrease in homeostasis-associated transcripts and an increase in transcripts associated with chronic neurodegenerative diseases, while astrocyte-associated genes showed an increase in transcripts associated with acute injury. Oligodendrocyte lineage cell genes showed a decrease in transcripts associated with both immature progenitors as well as myelinating oligodendrocytes. These data provide new insight into the mechanisms by which ethanol induces cerebellar neuropathology and alterations to the immune response in AUD.
Keywords: AUD, astrocyte, microglia, oligodendrocyte, neuroinflammation, transcriptomics
1. Introduction
Excessive alcohol consumption in adolescents and adults has significant societal impacts, with an estimated economic cost of $249 billion in the U.S. alone [1]. Studies have shown that alcohol misuse can lead to low academic achievement, an increased risk of suicide, and a lifetime struggle with addiction [2,3,4]. Furthermore, alcohol use disorder (AUD) is one of the most prevalent mental health disorders, with 15.7 million Americans aged 12 and older diagnosed [5,6], and is associated with many physical and psychiatric comorbidities [7,8]. Despite the known consequences of excess alcohol consumption, 29.7% of men and 22.2% of women were diagnosed with an AUD in 2019 [9]. AUD is associated with pathology to organ systems including the central nervous system (CNS). Animal models of AUD have been developed which simulate the behavioral abnormalities and neuropathologies associated with human AUD, thus allowing researchers to investigate the biological mechanisms associated with AUD [10]. Within the CNS, the cerebellum is responsible for coordinating motor movements, cognitive processing, and sensory discrimination. In individuals with AUD, these cerebellar functions are often disrupted, which may persist following abstinence from alcohol [11,12]. Alcohol can induce an immune response in the CNS termed neuroinflammation, which may result in neurodegeneration [13] and an increased risk of developing an AUD [14]. In adult rodents, the extent of alcohol-induced neuroinflammation can depend on the experimental paradigm of ethanol exposure utilized [15,16,17,18,19].
In the current study, we evaluated the effects of ethanol on the transcriptomic profile of adult mouse cerebella, utilizing a chronic plus binge ethanol exposure paradigm adapted from an alcoholic liver disease model developed by the Gao laboratory, in which liver injury and systemic inflammation were reported [20,21]. Using a top-down approach, we analyzed the effects of ethanol on global gene expression in the cerebellum. Our studies indicated that ethanol altered the expression of immune-related transcripts and pathways in the adult cerebellum, and may alter the function and phenotype of CNS glial cells. Thus, the current studies aid in advancing our understanding of the neuroinflammatory transcriptomic changes induced in AUD, unraveling potential targets for therapeutic strategies.
2. Materials and Methods
2.1. Animals
All animal use protocols were reviewed and approved by the University of Arkansas for Medical Sciences (UAMS), Institutional Animal Care and Use Committee (IACUC). Adult C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME, USA; stock #000664) and were housed in the UAMS Division of Laboratory Animal Medicine, where a breeding colony was established to produce experimental animals. Adult male mice aged 10–14 weeks and weighing ≥20 g were housed individually and were randomly separated into 2 experimental groups, ethanol (E) or vehicle control (C), (n = 5 mice per group). Solid food was removed from cages, while water was provided ad libitum for the duration of the study. On study days 1–5, both experimental groups of mice were allowed to acclimate to the Bio-Serv Rodent Liquid Diet, control formulation (Flemington, NJ, USA; #F1259SP) provided freely in a fresh tube each day just before the start of the dark cycle. Following acclimation, the ethanol group underwent ethanol ramping, in which mice received successive increases of the Bio-Serv ethanol formulation (#F1258SP) with either 1% (day 6), 2% (day 7), or 3% ethanol (day 8) diluted using 95% v/v ethanol (Acros, a part of Thermo Fisher Scientific, Waltham, MA, USA; #AC615110010). On study day 9, chronic ethanol administration began, in which the ethanol-treated mice received 4% ethanol for 10 days, followed by 5% ethanol for 7 days. Pair-feeding for the control group began on study day 10 (the second day of 4% ethanol administration), in which the control group was fed an equivalent volume of control diet to match the mean ethanol group consumption volume from the previous day. On the morning of study day 26, immediately following the start of the light cycle, the ethanol group underwent an acute binge administration of 5 g/kg of 31.5% ethanol (v/v) diluted from 95% v/v ethanol delivered in water via gavage. The control group received 45% (w/v) Maltose Dextrin (10 DE Food Grade #3585) diluted in water and delivered via gavage. At this time, the liquid diet was removed from all cages and standard food pellets were provided. 24 h following the ethanol binge administration, mice were euthanized and transcardially perfused with 1X PBS containing 5 U/mL heparin. Brains were removed and cerebella were micro-dissected into two halves along the midline and snap frozen in liquid nitrogen. Blood ethanol concentrations (BECs) from a separate set of animals were determined to be 230 (±59.7) mg/dL following 4% administration, 311.7 (±49.8) following 5% administration, and 718 (±6.9) mg/dL following bolus administration, as reported previously when using this model [22]. BECs were not measured at the time of tissue collection, though we suspect BECs were at or near 0 based upon preliminary studies using this model.
2.2. Isolation of RNA, RNA-Seq Library Preparation, and Sequencing
One whole cerebellar hemisphere from each experimental animal was homogenized using a B2X24B Bullet Blender and 0.5 mm glass beads, as described by the manufacturer (Next Advance, Troy, NY, USA). RNA was isolated using the RNeasy Lipid Tissue Mini Kit with on-column Dnase digestion using the Rnase-free Dnase Set (Qiagen, Valencia, CA, USA, Cat #74804 and #79254), as described previously [23]. RNA quantity was assessed using the Qubit 3.0 fluorometer with the Qubit Broad-Range RNA Assay Kit (Thermo Fisher Scientific), and an Agilent Fragment Analyzer with the Standard Sensitivity RNA Gel Kit (Agilent Technologies, Santa Clara, CA, USA) was used to ensure RNA quality. RNA-seq libraries were prepared using an Illumina TruSeq mRNA Library Prep Kit with TruSeq Unique Dual Indexed adapters (Illumina, San Diego, CA, USA), and were quantified with Qubit 1X dsDNA High-Sensitivity NGS Gel Kit (Thermo Fisher Scientific). KAPA Library Quantification (Roche, Basel, Switzerland) was used for further library characterization, and an Agilent Fragment Analyzer with the High-sensitivity NGS Gel Kit (Agilent) was used for determining fragment size. Library molarities were calculated followed by dilution and denaturation according to manufacturer’s specification for clustering. The control and ethanol-exposed animals were clustered on a high-output NextSeq 500 flow cell and paired-end sequenced with 150-cycle SBS kit for 2X75 reads (Illumina).
2.3. Bioinformatic Analysis
To identify significant differences in mRNA gene expression and global biological pathways associated with alterations of cerebellar genes between the control and ethanol treatment groups, raw RNA-sequence data (NCBI GEO accession GSE222445) were analyzed. RNA-seq reads were quality-checked, trimmed, and aligned to the GRCm39 reference genome (accession: GCA_000001635.9) using the Nextflow RNAseq pipeline, nf-core/rnaseq (version 3.4), available at DOI 10.5281/zenodo.1400710. The resulting gene counts were transformed to Log2 counts per million (CPM) [24]. Lowly expressed genes were filtered out, and libraries were normalized by trimmed means of M-values [25]. The Limma R package was used to calculate differential expression among genes [26]. Log2 fold change values were calculated for ethanol compared to control, and genes with an adjusted (adj.) p ≤ 0.05 were considered statistically significant.
Heat map and principal component analysis (PCA) plots were created from the processed differential gene expression data using R statistical software. The R-based EnhancedVolcano package was used to make the volcano plots [27]. Pathway and network analysis were conducted using the QIAGEN Ingenuity Pathway Analysis (IPA) software (QIAGEN Inc., Valencia, CA, USA, https://digitalinsights.qiagen.com/IPA, accessed on 22 July 2022 ) using the “Core Expression Analysis”. IPA analysis parameters were set with the “species” parameter as “mouse”, and the “tissues and cell lines” parameter as “cerebellum”, with gene cut offs of an adj. p ≤ 0.05 and Log2 fold change ≥0.5 or ≤−0.5.
To obtain a better understanding of the specific cellular processes and cell types of the cerebellum that are most sensitive to ethanol exposure, we extracted cell type-specific gene lists from publicly available single-cell RNA-seq (scRNA-seq) resources, which have been used previously to deduce the cell composition of bulk RNA-seq tissue [28]. Using this approach, we identified a total of 822 microglia-associated genes from scRNA-seq resources [29,30,31,32,33] (Supplemental Table S1A). We compared this list of microglia-associated genes to the list of genes significantly differentially regulated by ethanol (adj. p ≤ 0.05) in our dataset, which identified 151 microglia-associated genes whose expression was altered by ethanol (Table 1).
Table 1.
Uncategorized microglia-associated genes dysregulated by ethanol exposure in the cerebellum. Genes were identified by cross-referencing our significantly (adjusted p < 0.05) differentially regulated gene list with the 822 microglia-associated genes extracted from previous studies [identified in [29,30,31,32,33]] (Supplemental Table S1A) using R statistical software, which identified 151 genes associated with microglia.
| Symbol | LogFC | Adj. p | Symbol | LogFC | Adj. p | Symbol | LogFC | Adj. p | Symbol | LogFC | Adj. p | Symbol | LogFC | Adj. p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FOSB | 2.81 | 0.0081 | IFRD1 | 0.65 | 0.0001 | SLC25A5 | 0.27 | 0.0010 | CMTM6 | −0.21 | 0.0438 | PIK3CD | −0.50 | 0.0057 |
| GPX3 | 2.68 | 1.77 × 10−9 | ZFP36 | 0.62 | 0.0027 | CCNL1 | 0.27 | 0.0035 | MKNK1 | −0.22 | 0.0422 | CTSS | −0.51 | 0.0005 |
| CCL2 | 2.44 | 0.0015 | KLF4 | 0.60 | 0.0238 | FTL1 | 0.26 | 0.0021 | EDEM2 | −0.23 | 0.0235 | PLD4 | −0.52 | 0.0208 |
| CDKN1A | 2.31 | 0.0007 | ANXA3 | 0.58 | 0.0021 | TMSB4X | 0.26 | 0.0037 | DOCK10 | −0.23 | 0.0350 | KCTD12 | −0.53 | 1.79 × 10−5 |
| FCNA | 2.06 | 0.0028 | ARHGDIB | 0.54 | 0.0103 | PTBP1 | 0.23 | 0.0289 | RGS3 | −0.23 | 0.0465 | IFI203 | −0.54 | 0.0313 |
| MAFF | 1.94 | 0.0002 | IER3 | 0.50 | 0.0012 | MYLIP | 0.23 | 0.0321 | TLN2 | −0.24 | 0.0188 | COL27A1 | −0.54 | 0.0433 |
| CCL7 | 1.81 | 0.0025 | IER2 | 0.50 | 0.0318 | BRD2 | 0.23 | 0.0038 | SLC38A6 | −0.24 | 0.0467 | HPGDS | −0.60 | 0.0100 |
| C5AR1 | 1.53 | 0.0044 | PROS1 | 0.48 | 0.0116 | KLF6 | 0.23 | 0.0368 | PLXDC2 | −0.24 | 0.0134 | UNC93B1 | −0.60 | 0.0014 |
| GM3002 | 1.40 | 0.0405 | ICAM1 | 0.46 | 0.0449 | MCL1 | 0.21 | 0.0160 | RGL2 | −0.25 | 0.0089 | TREM2 | −0.62 | 0.0170 |
| MSR1 | 1.34 | 0.0221 | CFH | 0.45 | 0.0092 | PCF11 | 0.21 | 0.0071 | PPCDC | −0.25 | 0.0401 | ITGAM | −0.65 | 0.0010 |
| EVI2B | 1.25 | 0.0051 | LAIR1 | 0.45 | 0.0055 | CLTC | 0.21 | 0.0070 | SLC29A3 | −0.25 | 0.0314 | CCR5 | −0.67 | 0.0274 |
| LYVE1 | 1.22 | 0.0164 | DUSP6 | 0.44 | 0.0070 | CYFIP1 | 0.20 | 0.0136 | ZFP90 | −0.25 | 0.0257 | SELPLG | −0.67 | 0.0003 |
| UCP2 | 1.20 | 0.0088 | REL | 0.44 | 0.0343 | ZCCHC2 | 0.20 | 0.0245 | SLCO2B1 | −0.28 | 0.0484 | DSN1 | −0.68 | 0.0116 |
| CSRNP1 | 1.10 | 8.39 × 10−6 | RGS2 | 0.43 | 0.0281 | FMNL1 | 0.19 | 0.0425 | CAMK1 | −0.28 | 0.0040 | IRF7 | −0.70 | 0.0273 |
| APOC1 | 1.05 | 0.0009 | TSPO | 0.42 | 0.0433 | SERINC3 | 0.19 | 0.0467 | GPR155 | −0.28 | 0.0130 | APOBEC1 | −0.70 | 0.0296 |
| SPP1 | 1.05 | 0.0315 | ZFP36L2 | 0.41 | 0.0021 | IL16 | 0.18 | 0.0149 | TLR3 | −0.30 | 0.0436 | HK2 | −0.77 | 0.0023 |
| MERTK | 1.00 | 0.0348 | CD300A | 0.41 | 0.0117 | ARPC2 | 0.17 | 0.0203 | AKR1B10 | −0.30 | 0.0100 | IFI27L2A | −0.77 | 0.0403 |
| F13A1 | 0.98 | 0.0109 | SAT1 | 0.41 | 0.0007 | PCNA | 0.17 | 0.0350 | UBC | −0.31 | 0.0056 | FGD2 | −0.83 | 0.0048 |
| SERPINB8 | 0.97 | 0.0282 | 1700017B05RIK | 0.40 | 0.0163 | UBE2J1 | 0.17 | 0.0384 | AGO4 | −0.32 | 0.0367 | LY86 | −0.84 | 0.0002 |
| KLF10 | 0.95 | 0.0022 | COTL1 | 0.39 | 0.0018 | ELMO1 | 0.16 | 0.0220 | APH1C | −0.35 | 0.0282 | FCRLS | −0.85 | 0.0032 |
| ATF3 | 0.94 | 0.0077 | ATF4 | 0.39 | 0.0003 | SEMA4D | −0.16 | 0.0484 | EPB41L2 | −0.35 | 0.0016 | HPGD | −0.87 | 0.0004 |
| HSPA1A | 0.92 | 0.0054 | SRGN | 0.37 | 0.0237 | ASAH1 | −0.17 | 0.0333 | LPCAT2 | −0.35 | 0.0344 | KLHL6 | −0.95 | 0.0173 |
| ARHGAP27 | 0.83 | 0.0001 | ISYNA1 | 0.35 | 0.0247 | B2M | −0.17 | 0.0416 | ARHGAP11A | −0.37 | 0.0465 | SIGLECH | −0.98 | 0.0005 |
| SOCS3 | 0.81 | 0.0258 | H3F3B | 0.33 | 0.0072 | LY6E | −0.19 | 0.0276 | HEXB | −0.38 | 0.0003 | OAS2 | −0.98 | 0.0095 |
| GPNMB | 0.79 | 0.0039 | PPP1R15A | 0.31 | 0.0263 | TPP1 | −0.19 | 0.0097 | CSF1R | −0.42 | 0.0020 | P2RY12 | −1.10 | 0.0001 |
| PHYHD1 | 0.78 | 1.08 × 10−5 | ARL4C | 0.30 | 0.0029 | SGPL1 | −0.20 | 0.0388 | MPEG1 | −0.42 | 0.0088 | CD74 | −1.18 | 0.0001 |
| CD68 | 0.73 | 0.0096 | CCDC9 | 0.29 | 0.0047 | IL6ST | −0.20 | 0.0219 | GPR34 | −0.43 | 0.0433 | H2-AA | −1.55 | 0.0029 |
| EGR1 | 0.72 | 0.0028 | HERPUD1 | 0.28 | 0.0076 | PMP22 | −0.20 | 0.0479 | CRYL1 | −0.44 | 0.0130 | |||
| SPARC | 0.71 | 2.21 × 10−8 | SKI | 0.28 | 0.0104 | RRBP1 | −0.20 | 0.0274 | SALL1 | −0.45 | 0.0173 | |||
| C3AR1 | 0.69 | 0.0154 | SERPINF1 | 0.28 | 0.0375 | AXL | −0.21 | 0.0334 | RENBP | −0.46 | 0.0219 | |||
| SH2B2 | 0.68 | 0.0052 | PTPRJ | 0.27 | 0.0060 | COMMD8 | −0.21 | 0.0440 | P2RY13 | −0.48 | 0.0356 | |||
We were able to characterize 23 of the 151 genes as being either homeostatic or neurodegenerative (Table 2), as defined in previous studies [33,34,35,36,37] (Supplemental Table S1B,C).
Table 2.
Categorized microglia-associated genes dysregulated by ethanol exposure in the cerebellum. The microglia-associated genes identified in our data set in Table 1 with an adjusted p < 0.05 and Log2 fold change ≥ 0.25 or ≤ −0.25 were further categorized as being homeostatic or neurodegenerative, as defined by previous studies [identified in [35,36,37]].
| Homeostatic | LogFC | Adj. p | Neurodegenerative | LogFC | Adj. p |
|---|---|---|---|---|---|
| MERTK | 1.00 | 0.0348 | GPX3 | 2.68 | 1.77 × 10−9 |
| EGR1 | 0.72 | 0.0028 | CCL2 | 2.44 | 0.0015 |
| SLCO2B1 | −0.28 | 0.0484 | MSR1 | 1.34 | 0.0221 |
| HEXB | −0.38 | 0.0003 | SPP1 | 1.05 | 0.0315 |
| CSF1R | −0.42 | 0.0020 | GPNMB | 0.79 | 0.0039 |
| GPR34 | −0.43 | 0.0433 | CD68 | 0.73 | 0.0096 |
| SALL1 | −0.45 | 0.0173 | LAIR1 | 0.45 | 0.0055 |
| P2RY13 | −0.48 | 0.0356 | TREM2 | −0.62 | 0.0170 |
| KCTD12 | −0.53 | 1.79 × 10−5 | |||
| Hpgds | −0.60 | 0.0100 | |||
| CCR5 | −0.67 | 0.0274 | |||
| FGD2 | −0.83 | 0.0048 | |||
| FCRLS | −0.85 | 0.0032 | |||
| Siglech | −0.98 | 0.0005 | |||
| P2RY12 | −1.10 | 0.0001 |
To further evaluate the effects of ethanol on homeostatic versus neurodegenerative microglial phenotypes, we computed mean z-scores to compare control versus ethanol for the transcripts associated with these phenotypes. Since the goal was to determine relative gene expression changes in our dataset, i.e., to determine whether the genes are up- or down-regulated due to ethanol, the average z-score was computed. We calculated the average z-score across individual genes in our extracted microglia homeostatic and neurodegenerative-associated gene lists, and then averaged these individual gene z-scores within each sample. The average z-score of each sample in the homeostatic and neurodegenerative group was then evaluated using a two-tailed Student’s t test, with p ≤ 0.05 being considered statistically significant. R statistical software was used to conduct the Student’s t-test as well as construct the average z-score graphs.
Similar to microglia, we utilized scRNA-seq data to compose a list of 309 astrocyte-associated genes (Supplemental Table S2) [37]. From this list we identified 56 astrocyte-associated genes that were differentially expressed in response to ethanol in our current study. We then characterized these transcripts as being associated with an astrocyte phenotype common to acute injury, chronic neurodegenerative diseases, or pan-injury (Table 3), the last of which includes genes associated with both acute injury and chronic neurodegenerative disease phenotypes [37].
Table 3.
Categorized astrocyte-associated genes dysregulated by ethanol exposure in the cerebellum. Genes were identified by cross-referencing our significantly (adjusted p < 0.05) differentially regulated gene list with the list of 309 astrocyte-associated genes extracted from a previous study [identified in [38]] (Supplemental Table S2) using R statistical software. The astrocyte-associated genes identified in our dataset were then further categorized as being associated with acute injury, chronic neurodegenerative diseases, or pan-injury, as described in a previous study [38].
| Acute Injury | LogFC | Adj. p | Pan Astrocytic | LogFC | Adj. p | Chronic Neurodegenerative Diseases | LogFC | Adj. p | |
|---|---|---|---|---|---|---|---|---|---|
| RCAN2 | 0.40 | 0.0091 | UCP2 | 1.20 | 0.0088 | S1PR1 | −0.33 | 0.0006 | |
| Lrrc58 | 0.31 | 0.0036 | ATF3 | 0.94 | 0.0077 | ARSK | −0.33 | 0.0089 | |
| ARL4C | 0.30 | 0.0029 | GPNMB | 0.79 | 0.0039 | COBL | −0.47 | 0.0172 | |
| PRELP | 0.27 | 0.0368 | LGALS3 | 0.67 | 0.0282 | ||||
| YWHAZ | 0.26 | 0.0014 | ARHGDIB | 0.54 | 0.0103 | ||||
| DNTTIP2 | 0.24 | 0.0244 | RHOJ | 0.46 | 0.0117 | ||||
| CDC42SE1 | 0.23 | 0.0082 | PARP3 | 0.45 | 0.0065 | ||||
| HINT1 | 0.22 | 0.0040 | TIMP3 | 0.38 | 0.0216 | ||||
| CARS | 0.22 | 0.0079 | AHNAK | 0.33 | 0.0173 | ||||
| IARS | 0.21 | 0.0097 | PPARGC1A | 0.26 | 0.0276 | ||||
| ARNTL | 0.19 | 0.0240 | ELOVL2 | 0.25 | 0.0113 | ||||
| LRRC41 | 0.19 | 0.0461 | MCL1 | 0.21 | 0.0160 | ||||
| SSBP3 | 0.19 | 0.0202 | AHCYL1 | 0.16 | 0.0148 | ||||
| BRCC3 | 0.19 | 0.0288 | B2M | −0.17 | 0.0416 | ||||
| LRRC59 | 0.18 | 0.0391 | DST | −0.21 | 0.0280 | ||||
| UBE2F | 0.18 | 0.0219 | SQLE | −0.27 | 0.0246 | ||||
| FARSB | 0.16 | 0.0366 | APLN | −0.28 | 0.0433 | ||||
| CNBP | 0.14 | 0.0482 | PTPRD | −0.33 | 0.0006 | ||||
| SGPL1 | −0.20 | 0.0388 | FLOT1 | −0.33 | 0.0116 | ||||
| AXL | −0.21 | 0.0334 | NSDHL | −0.35 | 0.0137 | ||||
| LAP3 | −0.21 | 0.0321 | HMGCS1 | −0.43 | 0.0002 | ||||
| SGCB | −0.21 | 0.0213 | CTSS | −0.51 | 0.0005 | ||||
| RNF141 | −0.27 | 0.0039 | VIM | −0.51 | 1.91 × 10−5 | ||||
| SYNE1 | −0.30 | 0.0102 | IDI1 | −0.55 | 0.0009 | ||||
| POLD4 | −0.34 | 0.0375 | IFIT3 | −0.75 | 0.0360 | ||||
| PLIN2 | −0.38 | 0.0084 | |||||||
| IL33 | −0.91 | 0.0001 | |||||||
| IGSF1 | −0.92 | 0.0057 | |||||||
To test for statistical significance, the average z-scores of each gene in our extracted acute, chronic, and pan-injury astrocyte-associated gene lists were generated, and these individual gene z-scores were then averaged within each sample in a manner consistent with the microglia described above. The Student’s t-test and average z-score graphs were constructed using R statistical software. Due to the small number of chronic neurodegenerative disease astrocyte-associated genes (n = 3), no z-score graph was generated for this group.
For oligodendrocyte lineage-associated genes, we extracted gene lists for oligodendrocyte precursor cells (OPCs) (381 genes), committed oligodendrocyte precursor cells (COPs) (55 genes), newly formed oligodendrocytes (NFOL) (9 genes), myelin-forming oligodendrocytes (MFOL) (347 genes), and mature oligodendrocytes (MOL) (7 genes) from publicly available scRNA-seq studies [29,30,39] (Supplemental Table S3), in a manner consistent with microglia and astrocytes described above, to determine which genes were significantly differentially regulated by ethanol. From these lists, we identified 71 differentially expressed genes associated with OPCs, 12 genes associated with COPs, 2 genes associated with NFOL, 2 genes associated with MOL, and 108 genes associated with MFOL within our significantly differentially regulated dataset (Table 4).
Table 4.
Categorized oligodendrocyte lineage-associated genes dysregulated by ethanol exposure in the cerebellum. Genes were identified by cross-referencing our significantly (adjusted p < 0.05) differentially regulated gene list with the list of OPC, COP, NFOL, MFOL and MOL-associated genes [identified in [29,30,39]] (Supplemental Table S3) using R statistical software.
| OPC | LogFC | Adj. p | OPC | LogFC | Adj. p | OPC | LogFC | Adj. p | OPC | LogFC | Adj. p | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PTPRN | 1.03 | 0.0011 | GNG3 | 0.27 | 0.0053 | PRKCB | −0.17 | 0.0390 | LNX1 | −0.37 | 0.0017 | |||||||
| SERPINA3N | 0.98 | 0.0120 | DSCAM | 0.27 | 0.0173 | DNM3 | −0.18 | 0.0334 | RSU1 | −0.40 | 0.0007 | |||||||
| SMOX | 0.90 | 0.0001 | NMNAT2 | 0.26 | 0.0130 | DISP2 | −0.18 | 0.0349 | JAM2 | −0.41 | 0.0006 | |||||||
| GPNMB | 0.79 | 0.0039 | CXADR | 0.25 | 0.0102 | DDAH1 | −0.20 | 0.0476 | PHLDB1 | −0.42 | 0.0004 | |||||||
| SORCS1 | 0.60 | 0.0003 | ABHD17B | 0.25 | 0.0113 | PCDH9 | −0.22 | 0.0174 | LBH | −0.44 | 0.0002 | |||||||
| MIDN | 0.42 | 0.0088 | SCG5 | 0.25 | 0.0033 | PCDH10 | −0.23 | 0.0301 | RAMP1 | −0.45 | 0.0003 | |||||||
| TRIL | 0.39 | 0.0116 | CHPT1 | 0.24 | 0.0110 | OMG | −0.23 | 0.0191 | EDNRB | −0.47 | 0.0027 | |||||||
| HIP1 | 0.35 | 0.0003 | PHACTR3 | 0.24 | 0.0278 | SLC35F1 | −0.24 | 0.0275 | COBL | −0.47 | 0.0172 | |||||||
| KANK1 | 0.33 | 0.0160 | EHD3 | 0.23 | 0.0139 | SLC22A15 | −0.24 | 0.0188 | GLTP | −0.48 | 0.0006 | |||||||
| ITGAV | 0.33 | 0.0034 | DLGAP1 | 0.20 | 0.0124 | PCDH17 | −0.25 | 0.0235 | GJC3 | −0.48 | 0.0001 | |||||||
| CALY | 0.32 | 0.0021 | ADORA1 | 0.20 | 0.0151 | ADCYAP1R1 | −0.25 | 0.0029 | PTN | −0.52 | 0.0002 | |||||||
| GPT2 | 0.31 | 0.0014 | ZCCHC24 | 0.20 | 0.0245 | SVIL | −0.26 | 0.0391 | PLXNB3 | −0.52 | 0.0105 | |||||||
| CASKIN2 | 0.31 | 0.0163 | PTPRE | 0.20 | 0.0168 | KLHL5 | −0.27 | 0.0075 | MMP15 | −0.56 | 0.0239 | |||||||
| KCNK3 | 0.30 | 0.0130 | RAB31 | 0.19 | 0.0231 | GRIA4 | −0.29 | 0.0018 | RCN1 | −0.65 | 0.0103 | |||||||
| NCALD | 0.30 | 0.0041 | NELL2 | 0.19 | 0.0125 | SERINC5 | −0.30 | 0.0016 | RLBP1 | −0.78 | 0.0021 | |||||||
| LRRFIP1 | 0.29 | 0.0024 | GNPTG | 0.18 | 0.0202 | KLHL13 | −0.31 | 0.0113 | EMID1 | −0.84 | 0.0013 | |||||||
| CAV2 | 0.28 | 0.0473 | GAD1 | 0.15 | 0.0246 | CSPG5 | −0.34 | 0.0086 | PLLP | −1.11 | 0.0001 | |||||||
| SDC3 | 0.28 | 0.0411 | NOVA1 | −0.16 | 0.0402 | GNB4 | −0.35 | 0.0008 | ||||||||||
| COP | LogFC | Adj. p | NFOL | LogFC | Adj. p | |||||||||||||
| TIMP4 | 0.42 | 0.0001 | H2-AB1 | −1.38 | 0.0007 | |||||||||||||
| SEZ6L | 0.40 | 0.0005 | SEMA4D | −0.16 | 0.0484 | |||||||||||||
| SIRT2 | −0.16 | 0.0479 | ||||||||||||||||
| SLC44A1 | −0.18 | 0.0460 | ||||||||||||||||
| EDIL3 | −0.20 | 0.0247 | ||||||||||||||||
| S100B | −0.24 | 0.0080 | ||||||||||||||||
| BCAS1 | −0.28 | 0.0412 | ||||||||||||||||
| CNP | −0.30 | 0.0066 | ||||||||||||||||
| GPR17 | −0.33 | 0.0116 | ||||||||||||||||
| EPB41L2 | −0.35 | 0.0016 | ||||||||||||||||
| LIMS2 | −0.38 | 0.0468 | ||||||||||||||||
| ENPP6 | −0.53 | 0.0036 | ||||||||||||||||
| MFOL | LogFC | Adj. p | MFOL | LogFC | Adj. p | MFOL | LogFC | Adj. p | MFOL | LogFC | Adj. p | MOL | LogFC | Adj. p | ||||
| APOD | 1.66 | 0.0001 | LAP3 | −0.21 | 0.0321 | SEPTIN4 | −0.35 | 0.0005 | UGT8A | −1.16 | 0.0020 | NINJ2 | −1.88 | 0.0005 | ||||
| HSPA1A | 0.92 | 0.0054 | ATP8A1 | −0.21 | 0.0091 | ERMN | −0.37 | 0.0346 | SERPINB1A | −1.28 | 3.22 × 10−5 | KLK6 | −1.04 | 0.0016 | ||||
| ADIPOR2 | 0.90 | 0.0018 | SCCPDH | −0.21 | 0.0377 | MAG | −0.39 | 0.0346 | OPALIN | −2.33 | 6.07 × 10−7 | |||||||
| GLUL | 0.79 | 0.0010 | FGFR2 | −0.21 | 0.0362 | QDPR | −0.41 | 0.0029 | ||||||||||
| PIM3 | 0.64 | 0.0005 | FNBP1 | −0.21 | 0.0116 | PHLDB1 | −0.42 | 0.0004 | ||||||||||
| KLF13 | 0.53 | 0.0001 | CCP110 | −0.22 | 0.0142 | MAP6D1 | −0.43 | 0.0002 | ||||||||||
| HAPLN2 | 0.42 | 0.0267 | DIP2A | −0.22 | 0.0113 | CRYAB | −0.43 | 0.0445 | ||||||||||
| TUBB4A | 0.41 | 0.0036 | PCDH9 | −0.22 | 0.0174 | ABCA8A | −0.46 | 0.0122 | ||||||||||
| FTH1 | 0.39 | 0.0054 | TPST1 | −0.23 | 0.0279 | GNG11 | −0.46 | 0.0049 | ||||||||||
| KNDC1 | 0.39 | 0.0335 | DOCK10 | −0.23 | 0.0350 | NIPA1 | −0.47 | 0.0001 | ||||||||||
| SLC38A2 | 0.34 | 0.0003 | CNTN2 | −0.23 | 0.0218 | GLTP | −0.48 | 0.0006 | ||||||||||
| SLC20A2 | 0.30 | 0.0013 | TULP4 | −0.23 | 0.0022 | GPR37 | −0.48 | 0.0005 | ||||||||||
| CFL2 | 0.28 | 0.0040 | OMG | −0.23 | 0.0191 | GJC3 | −0.48 | 0.0001 | ||||||||||
| ZDHHC20 | 0.24 | 0.0249 | EPS15 | −0.24 | 0.0189 | CAR2 | −0.50 | 0.0010 | ||||||||||
| NUDT4 | 0.24 | 0.0047 | ARAP2 | −0.24 | 0.0130 | PRR5L | −0.50 | 0.0043 | ||||||||||
| LPGAT1 | 0.21 | 0.0097 | AATK | −0.25 | 0.0321 | ANO4 | −0.50 | 0.0010 | ||||||||||
| PAK1 | 0.21 | 0.0071 | SEMA6D | −0.25 | 0.0062 | ARSG | −0.52 | 0.0029 | ||||||||||
| TMOD2 | 0.20 | 0.0160 | KCNA6 | −0.27 | 0.0047 | PLXNB3 | −0.52 | 0.0105 | ||||||||||
| GPX4 | 0.20 | 0.0175 | GATM | −0.27 | 0.0091 | 1700047M11RIK | −0.53 | 0.0012 | ||||||||||
| PSAT1 | 0.19 | 0.0409 | BCAS1 | −0.28 | 0.0412 | LPAR1 | −0.54 | 0.0012 | ||||||||||
| PCNP | 0.18 | 0.0231 | S1PR5 | −0.29 | 0.0214 | TMEM88B | −0.56 | 0.0002 | ||||||||||
| CDC37L1 | 0.16 | 0.0424 | GRM3 | −0.29 | 0.0346 | CMTM5 | −0.59 | 0.0017 | ||||||||||
| ATP6AP2 | 0.16 | 0.0309 | EPHB1 | −0.29 | 0.0059 | FA2H | −0.67 | 0.0004 | ||||||||||
| DENND5A | −0.16 | 0.0239 | UNC5B | −0.29 | 0.0226 | ASPA | −0.67 | 0.0001 | ||||||||||
| ACOT7 | −0.17 | 0.0496 | TMEFF1 | −0.30 | 0.0304 | HHIP | −0.73 | 0.0033 | ||||||||||
| MYO6 | −0.17 | 0.0271 | SERINC5 | −0.30 | 0.0016 | TMEM125 | −0.75 | 0.0102 | ||||||||||
| SLC44A1 | −0.18 | 0.0460 | CNP | −0.30 | 0.0066 | SOX2OT | −0.85 | 0.0052 | ||||||||||
| SORT1 | −0.18 | 0.0127 | TTYH2 | −0.31 | 0.0053 | PPP1R14A | −0.86 | 0.0011 | ||||||||||
| DNM3 | −0.18 | 0.0334 | TPPP | −0.32 | 0.0026 | MOG | −0.86 | 0.0010 | ||||||||||
| ANK3 | −0.19 | 0.0130 | TRIM59 | −0.33 | 0.0334 | PDLIM2 | −0.87 | 0.0014 | ||||||||||
| YPEL2 | −0.20 | 0.0410 | REEP3 | −0.33 | 0.0022 | IL33 | −0.91 | 0.0001 | ||||||||||
| EDIL3 | −0.20 | 0.0247 | PTPRD | −0.33 | 0.0006 | PRR18 | −0.91 | 0.0003 | ||||||||||
| KCNJ10 | −0.20 | 0.0348 | PACS2 | −0.34 | 0.0008 | PLP1 | −1.07 | 5.01 × 10−7 | ||||||||||
| WNK1 | −0.20 | 0.0039 | DPY19L1 | −0.34 | 0.0012 | PLLP | −1.11 | 0.0001 | ||||||||||
| DST | −0.21 | 0.0280 | TSPAN2 | −0.35 | 0.0008 | GJC2 | −1.11 | 0.0043 | ||||||||||
We performed statistical analyses in a manner similar to the microglia and astrocytes above. Briefly, the average z-scores of each gene in our OPC, COP, and MFOL-associated gene lists were generated, and the individual gene z-scores were then averaged between each sample. The Student’s t-test and average z-score graphs were constructed using R statistical software. Due to the small number of NFOL and MOL-associated genes differentially regulated by ethanol, z-score graphs were not generated for these groups.
3. Results
3.1. Alcohol-Induced Differential Gene Expression in the Cerebellum
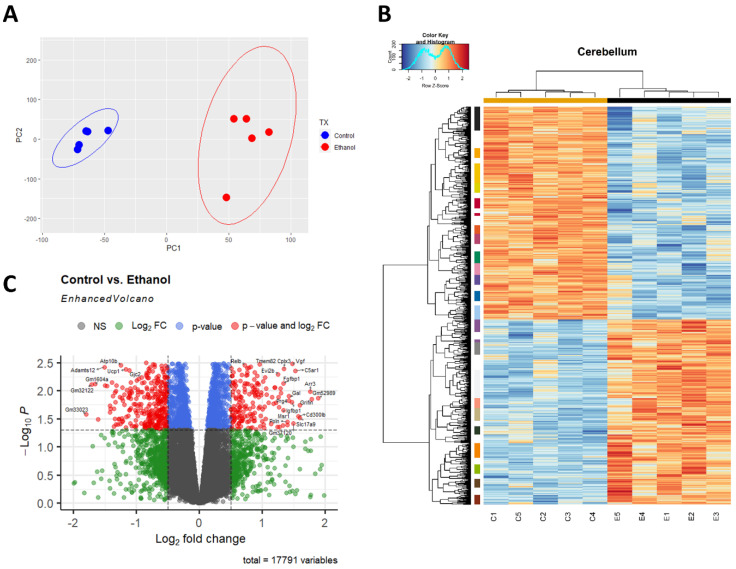
A principal component analysis (PCA) was performed to provide an overview of the transcriptomic changes that occurred in response to ethanol. PCA analysis demonstrated that gene transcripts correlating and anticorrelating to the first and second principal components could differentiate control animals from those exposed to ethanol. (Figure 1A). Hierarchical clustering analysis of significant genes was conducted using Pearson’s correlation, while controlling for false discovery rate adj. p ≤ 0.05 (Figure 1B). RNA-seq analysis identified 732 genes that were significantly differentially regulated (adj. p ≤ 0.05 and log2FC 0.5). Of these 732 genes, 269 were upregulated genes (36.75%) and 463 were downregulated genes (63.25%), (Figure 1C).
Figure 1.
Ethanol-induced differential gene expression in the cerebellum. Principle component analysis (PCA) of genes contributing to variance between ethanol (E) and control (C) in the cerebellum were analyzed using R statistical software (A). A heatmap and hierarchical clustering dendrogram of relative gene expression across samples was constructed using R statistical software for significantly (adjusted p < 0.05) altered genes. Red indicates positive z-scores (upregulation) and blue indicates negative z-scores (downregulation) (B). The R EnhancedVolcano package was utilized to construct a volcano plot displaying fold change versus adjusted p-value of all detected genes in the cerebellum. 732 of 17,791 total identified transcripts displayed an adjusted p < 0.05 and Log2 fold change ≥0.5 or ≤−0.5, shown in red (C). n = 5 males per treatment group E or C.
3.2. Pathway Analysis of the Alcohol-Induced Differentially Regulated Genes
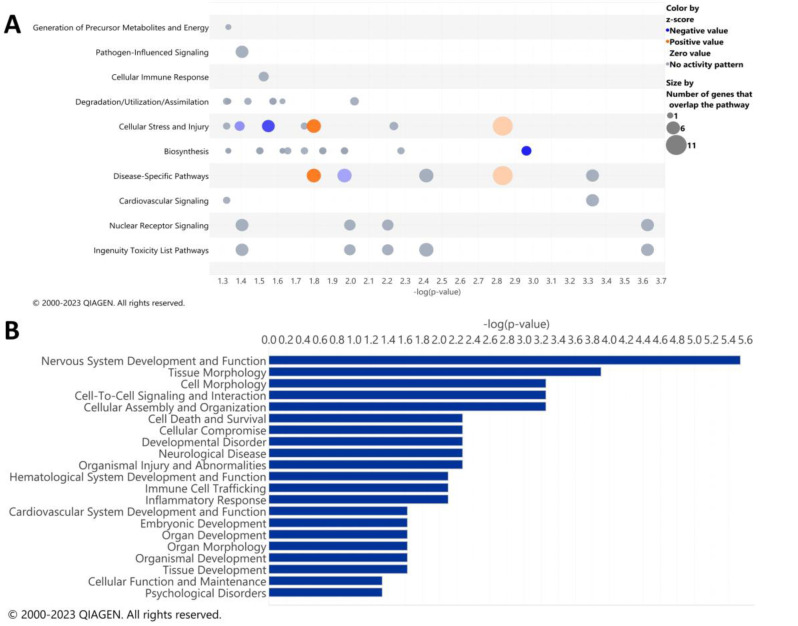
IPA analysis was performed to determine the specific pathways altered by ethanol in the cerebella of adult mice. The results of the top canonical pathways altered by ethanol exposure included those related to the generation of precursor metabolites and energy, pathogen-influenced signaling, cellular immune response, degradation/utilization/assimilation, cellular stress and injury, biosynthesis, disease-specific pathways, cardiovascular signaling, nuclear receptor signaling, and ingenuity toxicity list pathways (Figure 2A). A description of the pathway names, p-values, and molecules associated with each significantly altered pathway category is shown in Table 5. The top disease and biological function categories altered by ethanol exposure included nervous system development and function, tissue/cell morphology, cell-to-cell signaling and interaction, cell death and survival, cellular compromise, immune cell trafficking, and inflammatory response [−log(p.val) range = 5.5–2.1] (Figure 2B).
Figure 2.
Top canonical pathways and top diseases and biological functions in the cerebellum altered by ethanol exposure. Qiagen Ingenuity Pathway Analysis (IPA) software was utilized to assess the top canonical pathways (A) and the top diseases and biological functions (B) altered by ethanol exposure using the “cerebellum” selected analysis settings. All analyses were restricted to genes with an adjusted p < 0.05 and Log2 fold change ≥ 0.5 or ≤−0.5. n = 5 males per treatment group E or C.
Table 5.
Tabular descriptions of the top canonical pathway categories, including pathway names, p-values, and indicated molecules. Qiagen Ingenuity Pathway Analysis (IPA) software was utilized to assess the top canonical pathways altered by ethanol exposure using the “cerebellum” selected analysis settings. All analyses were restricted to genes with an adjusted p < 0.05 and Log2 fold change ≥0.5 or ≤−0.5.
| Pathway Category | Pathway Name | p-Value | Molecules |
|---|---|---|---|
| Generation of precursor metabolites and energy | Glycerol-3-phosphate shuttle | 0.0469 | GPD1 |
| Pathogen-influenced signaling | LPS/IL-1 mediated inhibition of RXR function | 0.0400 | CHST7, GSTM5, IL33, RARA, SMOX, SREBF1 |
| Cellular immune response | Granulocyte adhesion and diapedesis | 0.0303 | C5AR1, IL33, SDC4, SELPLG |
| Degradation/utilization/assimilation | Tryptophan degradation X | 0.0481 | AKR1B10, SMOX |
| Glycerol degradation I | 0.0469 | GPD1 | |
| Dopamine degradation | 0.0368 | SMOX, Sult1a1 | |
| Acetone degradation I (to Methylglyoxal) | 0.0268 | AKR1B10, CYP51A1 | |
| Spermine and spermidine degradation I | 0.0237 | SMOX | |
| Cellular stress and injury | Intrinsic prothrombin activation pathway | 0.0481 | COL5A3, KLK6 |
| GP6 signaling pathway | 0.0388 | COL16A1, COL27A1, COL5A1, COL5A3 | |
| Wound-healing signaling pathway | 0.0288 | COL16A1, COL27A1, COL5A1, COL5A3, IL33, VIM | |
| Coagulation system | 0.0181 | F3, VWF | |
| Osteoarthritis pathway | 0.0163 | ANXA2, FGFR3, GREM1, HES1, HTRA1, SDC4, SPP1 | |
| Apelin liver signaling pathway | 0.0059 | AGT, COL5A3, EDN1 | |
| Pulomary fibrosis idiopathic signaling pathway | 0.0015 | CCN2, COL16A1, COL27A1, COL5A1, COL5A3, EDN1, EGR1, FGFR3, HES1, LPAR1, VIM | |
| Biosynthesis | Trans, trans-faresyl diphosphate biosynthesis | 0.0469 | IDI1 |
| Cholesterol biosynthesis III (via desmosterol) | 0.0316 | CYP51A1, MSMO1 | |
| Glutamine biosynthesis I | 0.0237 | GLUL | |
| Superpathway of citrulline metabolism | 0.0223 | ASL, PRODH | |
| Γ-linolenate biosynthesis II | 0.0181 | FADS1, FADS2 | |
| Superpathway of geranylgeranyldiphosphate biosynthesis I (via mevalonate) | 0.0143 | ACAT2, IDI1 | |
| Mevalonate pathway I | 0.0109 | ACAT2, IDI1 | |
| Zymosterol biosynthesis | 0.0054 | CYP51A1, MSMO1 | |
| Superpathway of cholesterol biosynthesis | 0.0011 | ACAT2, CYP51A1, IDI1, MSMO1 | |
| Disease-specific pathway | Osteoarthritis pathway | 0.0163 | ANXA2, FGFR3, GREM1, HES1, HTRA1, SDC4, SPP1 |
| Pathogen-induced cytokine storm signaling pathway | 0.0111 | COL16A1, COL27A1, COL5A1, COL5A3, DHX58, IL33, SOCS3 | |
| Hepatic fibrosis/hepatic stellate cell activation | 0.0040 | AGT, CCN2, COL16A1, COL27A1, COL5A1, COL5A3, EDN1 | |
| Pulomary fibrosis idiopathic signaling pathway | 0.0015 | CCN2, COL16A1, COL27A1, COL5A1, COL5A3, EDN1, EGR1, FGFR3, HES1, LPAR1, VIM | |
| Atherosclerosis signaling | 0.0005 | APOD, COL5A3, F3, IL33, SELPLG, TNFRSF12A | |
| Cardiovascular signaling | Intrinsic prothrombin activation pathway | 0.0481 | COL5A3, KLK6 |
| Atherosclerosis signaling | 0.0005 | APOD, COL5A3, F3, IL33, SELPLG, TNFRSF12A | |
| Nuclear receptor signaling | LPS/IL-1 mediated inhibition of RXR function | 0.0400 | CHST7, GSTM5, IL33, RARA, SMOX, SREBF1 |
| LXR/RXR activation | 0.0103 | AGT, APOD, CYP51A1, IL33, SREBF1 | |
| FXR/RXR activation | 0.0064 | AGT, APOD, IL33, RARA, SREBF1 | |
| VDR/RXR activation | 0.0002 | CDKN1A, HES1, IGFBP1, KLF4, KLK6, SPP1 | |
| Ingenuity toxicity list pathways | LPS/IL-1 mediated inhibition of RXR function | 0.0400 | CHST7, GSTM5, IL33, RARA, SMOX, SREBF1 |
| LXR/RXR activation | 0.0103 | AGT, APOD, CYP51A1, IL33, SREBF1 | |
| FXR/RXR activation | 0.0064 | AGT, APOD, IL33, RARA, SREBF1 | |
| Hepatic fibrosis/hepatic stellate cell activation | 0.0040 | AGT, CCN2, COL16A1, COL27A1, COL5A1, COL5A3, EDN1 | |
| VDR/RXR activation | 0.0002 | CDKN1A, HES1, IGFBP1, KLF4, KLK6, SPP1 |
The diseases and biological function annotations that correlate to the diseases and biological functions categories, as shown in Figure 2B, are myelination (p.val = 2.88 × 10−6 ) or demyelination (p.val = 0.0053) of the cerebellum; quantity (p.val = 0.000125) or coupling (p.val = 0.000556) of oligodendrocytes; thickness of myelin sheath (p.val = 0.000556); quantity of cells (p.val = 0.00783); activation of microglia (p.val = 0.00783); permeability of blood–brain barrier (p.val = 0.0236); and astrocytosis of cerebella (p.val = 0.0467), (Table 6). These results suggest that in the cerebellum, ethanol alters biological functions that pertain to alterations in the formation of myelin, along with possible microglia and astrocyte phenotypic changes.
Table 6.
Tabular descriptions of the disease and biological function categories, including annotation, p-value, and indicated molecules. Qiagen Ingenuity Pathway Analysis (IPA) software was utilized to assess the top diseases and biological functions altered by ethanol exposure using the “cerebellum” selected analysis settings. All analyses were restricted to genes with an adjusted p < 0.05 and Log2 fold change ≥ 0.5 or ≤−0.5.
| Categories | Disease or Function Annotation | p-Value | Molecules |
|---|---|---|---|
| Nervous system development and function | Myelination | 2.88 × 10−6 | ASPA, FGFR3, GJB6, GJC2, HPGDS |
| Nervous system development and function, tissue Morphology | Quantity of oligodendrocytes | 0.000125 | FGFR3, GJB6, GJC2 |
| Cell-to-cell signaling and interaction | Coupling of oligodendrocytes | 0.000556 | GJB6, GJC2 |
| Cell morphology, cellular assembly and organization, nervous system development and function, tissue morphology | Thickness of myelin sheath | 0.000556 | GJB6, GJC2 |
| Cell-to-cell signaling and interaction | Coupling of astrocytes | 0.000556 | GJB6, GJC2 |
| Cellular assembly and organization | Formation of vacuole | 0.00164 | GJB6, GJC2 |
| Developmental disorder, nervous system development and function, neurological disease, organismal injury and abnormalities | Demyelination of cerebellum | 0.0053 | ASPA, HPGDS |
| Cell death and survival, cellular compromise, neurological disease, organismal injury and abnormalities, tissue morphology | Neurodegeneration of axons | 0.0053 | ASPA, SPTSSB |
| Tissue morphology | Quantity of cells | 0.00738 | ARSG, ASPA, FGFR3, GJB6, GJC2, NRN1 |
| Cell-to-cell signaling and interaction, hematological system development and function, immune cell trafficking, inflammatory response, nervous system development and function | Activation of microglia | 0.00783 | GJB6, GJC2 |
| Nervous system development and function | Morphology of nervous system | 0.011 | ARSG, FA2H, GJB6, GJC2, MERTK, PLP1, RARA, TBATA, UGT8, ZIC4 |
| Nervous system development and function, tissue morphology | Morphology of nervous tissue | 0.0126 | ARSG, FA2H, GJB6, GJC2, PLP1, TBATA, UGT8 |
| Cellular compromise, neurological disease, organismal injury and abnormalities | Damage of axons | 0.0236 | SOCS3 |
| Cell-to-cell signaling and interaction, nervous system development and function | Synaptic transmission of Bergmann glia | 0.0236 | SLC1A6 |
| Embryonic development, nervous system development and function, organ development, organismal development, tissue development | Delay in myelination of cerebellum | 0.0236 | FGFR3 |
| Cardiovascular system development and function, nervous system development and function, organ morphology, tissue morphology | Permeability of blood–brain barrier | 0.0236 | MOG |
| Nervous system development and function, neurological disease, organismal injury and abnormalities | Abnormal morphology of nervous system | 0.0314 | ARSG, FA2H, MERTK, PLP1, RARA, TBATA, UGT8, ZIC4 |
| Cellular assembly and organization, cellular function and maintenance, nervous system development and function, tissue morphology | Quantity of dendrites | 0.0467 | NRN1 |
| Neurological disease, organismal injury and abnormalities, psychological disorders | Spongy degeneration of central nervous system of white matter | 0.0467 | ASPA |
| Neurological disease, organismal injury and abnormalities | Astrocytosis of cerebellum | 0.0467 | HPGDS |
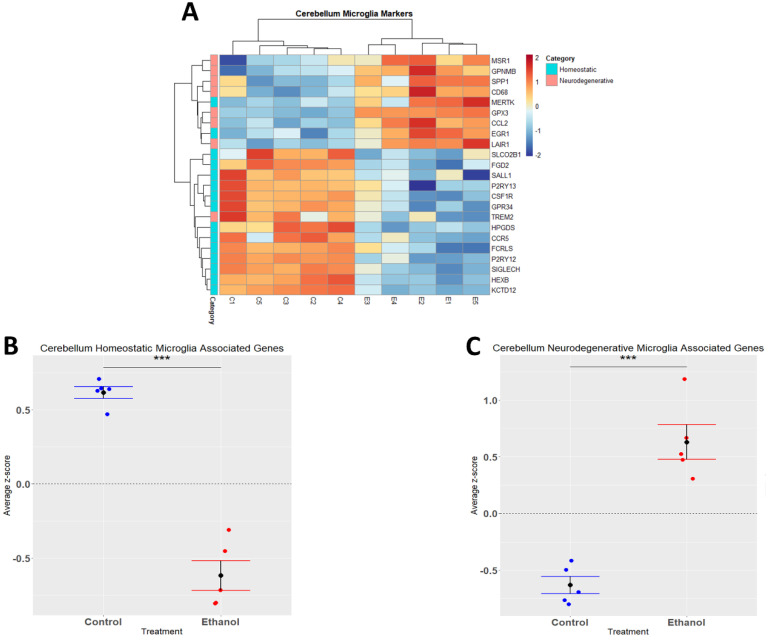
3.3. Alcohol Suppresses Microglia Homeostatic Genes while Increasing the Expression of Microglia Neurodegenerative-Associated Genes
Alcohol has been demonstrated to induce neuroinflammation in both humans and rodents which may include microglial activation, characterized by shortening and thickening of processes, along with the secretion of proinflammatory cytokines and chemokines that may contribute to neuropathology [19,40,41]. We performed hierarchical clustering analysis on homeostatic and neurodegenerative disease microglia-associated genes that were differentially expressed (adj. p ≤ 0.05) in response to ethanol (Figure 3A). A Student’s t-test comparing the average z-scores across all relevant genes indicated that ethanol caused an overall significant downregulation of microglia homeostatic genes (p.val = 3.191 × 10−6) (Figure 3B, Table 2) and an overall significant upregulation of microglia genes associated with neurodegenerative diseases (p.val = 7.786 × 10−5) (Figure 3C, Table 2). Collectively, these data suggest that ethanol may alter the microglial phenotype from a homeostatic and protective phenotype to a more activated phenotype observed in neurodegenerative diseases.
Figure 3.
Microglia-associated genes altered by ethanol exposure in the cerebellum. R statistical software was utilized to construct a heatmap and hierarchical clustering dendrogram of relative gene expression across samples for significantly (adjusted p < 0.05) altered and categorized microglia-associated genes as detailed in Methods. Red indicates positive z-scores (upregulation) and blue indicates negative z-scores (downregulation) (A). Individual genes were z-scored across samples, followed by calculation of average z-score for each treatment group which was used for testing statistical significance in R with Student’s t-test. Quantification by average z-score of homeostatic microglia-associated genes (B) and neurodegenerative microglia-associated genes (C). n = 5 males per treatment group E or C; *** p < 0.001.
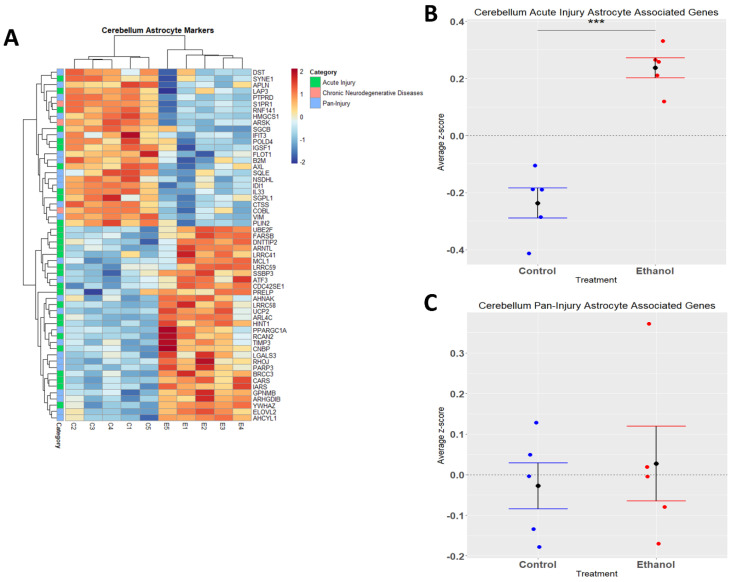
3.4. Astrocytes Undergo a Phenotypic Switch following Chronic plus Binge-like Alcohol Exposure
Astrocytes are one of the most abundant cell types in the CNS and play a critical role in regulating CNS functions in health and disease by maintaining homeostasis, providing energy to neurons, regulating synapse development and plasticity, modulating blood-brain-barrier integrity, and controlling neurological function and behavior [42,43,44,45,46]. Similarly to microglia, astrocytes play a role in CNS inflammation [47,48], and ethanol has been demonstrated to trigger an immune response in astrocytes [49,50]. In the current study, we performed hierarchical clustering analysis on acute injury, chronic neurodegenerative, and pan-injury astrocyte-associated genes that were differentially expressed (adj. p ≤ 0.05) in response to ethanol (Figure 4A). A Student’s t-test comparing the average z-scores across all relevant genes indicated that ethanol caused an overall significant increase in astrocyte genes related to acute injury (p.val = 7.085 × 10−5) (Figure 4B, Table 3) and an almost even number of up- and down-regulated genes (12 up vs. 13 down) pertaining to pan-injury (p.val = 0.6266) (Figure 4C, Table 3). Ethanol only altered the expression of three genes associated with the chronic neurodegenerative disease category (Table 3), thus the effect of ethanol on this small number of genes was not statistically evaluated. These data suggest that alcohol-induced transcriptomic changes in astrocytes are consistent with an acute injury phenotype.
Figure 4.
Astrocyte-associated genes altered by ethanol exposure in the cerebellum. R statistical software was utilized to construct a heatmap and hierarchical clustering dendrogram of relative gene expression across samples for significantly (adjusted p < 0.05) altered and categorized astrocyte-associated genes, as detailed in Methods. Red indicates positive z-scores (upregulation) and blue indicates negative z-scores (downregulation) (A). Individual genes were z-scored across samples, followed by calculation of the average z-score for each treatment group, which was used for testing statistical significance in R with Student’s t-test. Quantification by average z-score of acute injury astrocyte-associated genes (B) and pan-injury astrocyte-associated genes (C). Due to the small number of chronic neurodegenerative injury astrocyte-associated genes, no z-score graph was generated for this group; however, this group is further characterized in Table 3. n = 5 males per treatment group E or C; *** p < 0.001.
3.5. Oligodendrocyte Lineage Cells Are Depleted upon Chronic plus Binge-like Alcohol Exposure
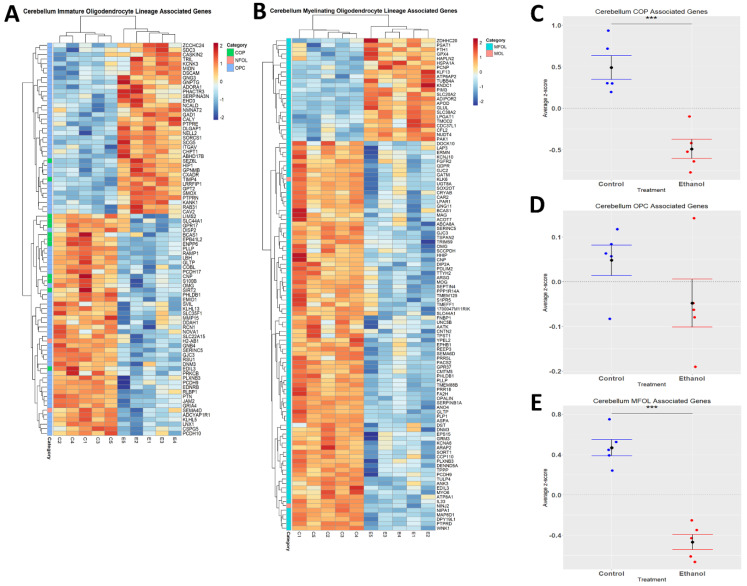
Ethanol has been demonstrated to alter myelination in adult humans and rodents [51,52]. We performed hierarchical clustering analysis on genes associated with distinct oligodendrocyte lineages (immature and myelinating) whose expression was altered by ethanol (Figure 5A,B). Evaluation of the effects of ethanol on immature oligodendrocyte lineages indicated that ethanol significantly decreased the expression of genes associated with COPs (p.val = 0.0006784) (Figure 5C, Table 4), and that ethanol skewed toward decreasing the expression of genes associated with OPCs (p.val = 0.1702) (Figure 5D, Table 4). For the myelinating oligodendrocyte lineage cells, ethanol significantly decreased the expression of genes associated with MFOLs (p.val = 2.905 × 10−05) (Figure 5E, Table 4). NFOL and MOL groups only contained two differentially expressed genes; therefore, statistical significance was not evaluated for these categories (Table 4). These results suggest that ethanol effects both immature and myelinating oligodendrocyte lineage cells, which could potentially lead to altered myelination.
Figure 5.
Alterations in oligodendrocyte lineage-associated genes by ethanol exposure in the cerebellum. R statistical software was utilized to construct a heatmap and hierarchical clustering dendrogram of relative gene expression across samples for significantly (adjusted p < 0.05) altered and categorized oligodendrocyte lineage-associated genes as detailed in Methods: immature oligodendrocyte lineage-associated genes (A) and myelinating oligodendrocyte lineage-associated genes (B) Red indicates positive z-scores (upregulation) and blue indicates negative z-scores (downregulation) (A,B). Individual genes were z-scored across samples, followed by calculation of average z-score for each treatment group, which was used for testing statistical significance in R with Student’s t-test. Quantification by average z-score of COP-associated genes (C), OPC-associated genes (D), and MFOL-associated genes in the cerebellum (E). Due to the small number of NFOL and MOL-associated genes, no z-score graph was generated for this group; however, this group is further characterized in Table 4. Abbreviations: OPC, oligodendrocyte precursor cell; COP, committed oligodendrocyte precursor; MFOL, myelin-forming oligodendrocyte; NFOL, newly formed oligodendrocyte; MOL, mature oligodendrocyte. n = 5 males per treatment group E or C; *** p < 0.001.
4. Discussion
Pathway analysis indicated that ethanol had significant effects on immune processes in the cerebella of adult mice. In addition, these analyses suggested that ethanol may alter the phenotype and function of glial cells including microglia, astrocytes, and oligodendrocyte lineage cells. We and others have previously demonstrated that ethanol induces neuroinflammation in adult rodents. However, the amount of neuroinflammation varies depending on the ethanol administration paradigm. For example, acute 4-day ethanol exposure did not alter the expression of pro-inflammatory molecules, although microglial activation was observed [17,53]. Following 10-day ethanol exposure, increased expression of pro-inflammatory molecules was observed, although it was somewhat modest [16,18,19]. Chronic ethanol exposure over a period of 3–5 months resulted in more robust neuroinflammation [15,49,54,55]. Using a variation of the same model as the current study, in which gene expression in both male and female mice was evaluated in control, ethanol, and ethanol + pioglitazone experimental groups, we have previously demonstrated robust neuroinflammation following chronic plus binge exposure to ethanol in less than one month [22]. This model is similar to an alcoholic liver disease model used previously by the Gao laboratory, in which they showed systemic inflammation and liver injury [20,21]. At this point, it is unclear in our studies whether ethanol induces CNS inflammation directly or indirectly through ethanol induced inflammation outside of the CNS. In order to begin to understand the possible mechanisms by which ethanol induces neuroinflammation in this chronic plus binge model of AUD, we have treated a unique set of male mice for the purpose of RNAseq analysis in the current study. We acknowledge that the use of only male mice is a limitation of the current study. Furthermore, some of the pathways identified in the current study only contain 1 or 2 genes, and some genes are represented in multiple pathways. Thus, we have exercised caution to not overinterpret the results.
We evaluated the transcriptomic data to identify immune-regulated genes whose expression was most strongly induced by ethanol, which included FOSB, CCL2, CCL7, C5AR1, SPP1, CD68, SOCS3, C3AR1, and KLF4. The most highly upregulated gene is FOSB, which encodes a transcription factor that dimerizes with Jun protein to form AP-1 and plays a critical role in alcohol and drug addiction [56]. Alcohol increases the expression of FOSB in the mesocorticolimbic system, which is believed to contribute to alcohol use disorder [57,58]. Furthermore, ethanol was demonstrated to alter synaptic plasticity and epigenetic alterations in the FOSB promoter, resulting in increased FOSB expression in the medial prefrontal cortex in wild-type but not TLR4 deficient mice. Since ethanol is believed to activate TLR4, resulting in downstream immune signaling [59], a role of ethanol-induced neuroinflammation is suggested in these processes. FOSB has also been demonstrated to contribute to excitotoxic microglial activation through regulation of complement C5a receptors in these cells [60]. Interestingly ethanol strongly increased the expression of complement C5AR1 and C3AR1 in our RNA-Seq studies. C5AR1 expression is increased in the liver of patients with alcoholic hepatitis [61], and is believed to contribute to alcohol-induced inflammation and liver injury [62,63]. Additionally, ethanol induces the expression of complement receptors including C3AR1 expression in microglia, resulting in altered phagocytosis [64]. We previously demonstrated that ethanol induces the expression of the chemokine CCL2 or MCP-1 following acute ethanol exposure in adult rodents [65], as well as in animal models of fetal alcohol spectrum disorders (FASD) [66]. It is interesting that in the current study, ethanol induced the expression of CCL2 as well the related chemokine CCL7 or MCP-3 in this chronic plus binge model. It should also be noted that transcriptomic changes were only evaluated at one timepoint, 24 h after the final ethanol exposure. Future studies may wish to evaluate transcriptomic changes at different times following the final ethanol exposure. It is also noteworthy that the other immune-related molecules we identified previously in this model were not indicated in the current study; this may be due to less sensitivity and smaller “n”, both of which are limitations that come with RNAseq when compared to quantitative real-time PCR [22].
Microglia are capable of responding to signals, resulting in activation and an altered phenotype. Our IPA analysis indicated that ethanol treatment resulted in microgliosis or microglial activation in the cerebellum. Upon activation, microglia have traditionally been hypothesized to undergo classical activation, resulting in a M1 pro-inflammatory phenotype, or alternative activation, resulting in an M2 anti-inflammatory or protective phenotype [67,68]. However, more recently it has become clear that microglial phenotypes are complex, and cannot be defined or categorized effectively using this simple binary system [69]. One recent nomenclature to distinguish microglial phenotype focuses on homeostatic versus neurodegenerative disease phenotypes. Under homeostatic conditions, microglia have a homeostatic phenotype, described by playing a role in synaptic plasticity and synaptogenesis, trophic support, chemotaxis and immune cell recruitment, and neurogenesis [37]. During insult to the CNS, microglia commonly lose their homeostatic signature and assume a chronic inflammatory signature [70,71,72]. Evaluation of the phenotype of microglia in a variety of neurodegenerative diseases have resulted in the identification of a common neurodegenerative disease-associated microglia phenotype [34,37,71,73]. In the current study, ethanol induced a microglia phenotypic switch in the cerebellum. This phenotypic switch was similar to that observed in neurodegenerative diseases, with a downregulation of homeostatic signature genes and an upregulation of neurodegenerative signature genes.
Astrocytes, like microglia, are capable of functioning in the innate immune response in the CNS. Once astrocytes are activated, commonly referred to as astrogliosis/astrocytosis, they produce cytokines and chemokines, nitric oxide, and other reactive oxygen species as part of an inflammatory response [74], Our IPA analysis indicated that ethanol treatment resulted in “astrocytosis”. Astrocytes were classically defined to respond to various stimuli to become reactive A1 astrocytes (neurotoxic or reactive A2 astrocytes) which are protective and neurotrophic [75,76]. However, as with microglia, this binary system of classifying reactive astrocytes appears inadequate to fully define and distinguish astrocyte phenotypes. More recently, Serrano-Pozo and colleagues performed a meta-analysis of mouse transcriptomic studies which resulted in a nomenclature that classified reactive astrocytes as being consistent with acute injury, chronic neurodegeneration, or pan-injury reactive astrocytes which exhibited characteristics of both acute injury and chronic neurodegenerative phenotypes [38]. In the current study, we determined that ethanol induced changes consistent with an acute injury astrocyte phenotype. Interestingly, LPS was previously shown to trigger an acute injury astrocyte phenotype [38]. ethanol has also been shown to activate TLR4 receptors, suggesting that ethanol-mediated neuroinflammation could occur in response to recruitment of TLR4 during alcohol use/abuse [77,78,79]. Therefore, we speculate that in this model of AUD, in the cerebellum, ethanol induces an acute injury astrocytic phenotype through the activation of TLR4, subsequently inducing an immune response.
Oligodendrocytes are responsible for forming a myelin sheath around axons of neurons in the CNS, facilitating the efficient propagation of action potentials [80]. OPCs are produced during embryogenesis, and migrate to their functional location wherein they differentiate into mature myelinating oligodendrocytes. Most myelination occurs at later stages of CNS development but can occur throughout life [81]. Ethanol has profound effects on the developing CNS and is believed to significantly contribute to the pathology associated with FASD, at least in part by altering myelination [82]. Ethanol also alters myelination in adults with AUD [83,84]. Ethanol is highly toxic to oligodendrocyte lineage cells, with OPCs being particularly susceptible [85,86]. Alcohol exposure is known to disrupt OPC differentiation and survival by decreasing the expression of platelet-derived growth factor receptor α (PDGFRα), a molecule crucial for differentiation of OPCs into mature oligodendrocytes [87]. In the current study, we found that adult chronic plus binge-like alcohol exposure depletes the expression of genes associated with both immature oligodendrocyte precursor cells as well as myelinating oligodendrocytes. Future studies are needed to determine the mechanism by which ethanol effects oligodendrocyte lineage cells and myelination in AUD.
5. Conclusions
The current study demonstrates that ethanol alters the transcriptomic profile in the adult cerebellum in a chronic plus binge model of AUD. The pathways altered by ethanol included those involved in immune response. Ethanol caused a shift in the expression of microglial-associated genes, with a decrease in homeostatic and an increase in chronic neurodegenerative-associated transcripts. Ethanol also increased the expression of astrocyte-associated genes common to acute injury. Finally, ethanol decreased the expression of genes associated with immature oligodendrocyte progenitor cells, as well as myelinating oligodendrocytes. These results provide clues about the mechanisms by which ethanol induces neuroinflammation and altered glial function in AUD.
Acknowledgments
RNA sequencing was performed by the UAMS Genomics Core which is supported by the Winthrop P. Rockefeller Cancer Institute, University of Arkansas for Medical Sciences.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/2073-4409/12/5/745/s1. Table S1A. Microglia associated genes [identified in [29,30,31,32,33]], Table S1B. Microglia homeostatic genes [identified in [35] and [37]], Table S1C. Common microglia genes affected during disease states [identified in [35,36,37]], Table S2. Categorized astrocyte associated genes [identified in [38]], Table S3. Categorized oligodendrocyte associated genes [identified in [29,30,39]].
Author Contributions
All authors had access to the data for the study, made substantial contributions to the manuscript, approved the submitted version of the manuscript, and take responsibility for the accuracy and integrity of the data. Conceptualization, P.D.D., C.J.M.K. and R.C.M.; Writing—Original Draft, P.D.D., K.N.H. and J.C.D., Writing—Review and Editing, K.N.H., M.R.P., J.C.D., T.M.R., C.J.M.K., R.C.M. and P.D.D.; Investigation, K.N.H., M.R.P., J.C.D. and T.M.R.; Formal Analysis, K.N.H., M.R.P. and J.C.D.; Visualization, K.N.H., M.R.P., J.C.D. and T.M.R.; Supervision, P.D.D. and R.C.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
This study was conducted according to the guidelines set forth by the University of Arkansas for Medical Sciences (UAMS) Office of Laboratory Animal Welfare and was approved by the UAMS Institutional Animal Care and Use Committee (IACUC), on 19 July 2021 (IACUC Animal Use Protocol (AUP), File #4120).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [88] and are accessible through GEO Series accession number GSE222445 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE222445, accessed on 24 February 2023).
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the National Institutes of Health: National Institute on Alcohol Abuse and Alcoholism [Grant/Award Numbers: R01 AA024695 (PDD), R01 AA026665 (PDD), R01 AA027111(PDD), F30 AA027698 (MRP)].
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Sacks J.J., Gonzales K.R., Bouchery E.E., Tomedi L.E., Brewer R.D. 2010 National and State Costs of Excessive Alcohol Consumption. Am. J. Prev. Med. 2015;49:e73–e79. doi: 10.1016/j.amepre.2015.05.031. [DOI] [PubMed] [Google Scholar]
- 2.Marshall E.J. Adolescent alcohol use: Risks and consequences. Alcohol Alcohol. 2014;49:160–164. doi: 10.1093/alcalc/agt180. [DOI] [PubMed] [Google Scholar]
- 3.Center for Behavioral Health Statistics and Quality . 2016 National Survey on Drug Use and Health: Detailed Tables. Substance Abuse and Mental Health Services Administration; Rockville, MD, USA: 2017. [Google Scholar]
- 4.White A., Hingson R. The burden of alcohol use: Excessive alcohol consumption and related consequences among college students. Alcohol Res. 2013;35:201–218. [PMC free article] [PubMed] [Google Scholar]
- 5.Grant B.F., Stinson F.S., Dawson D.A., Chou S.P., Dufour M.C., Compton W., Roger P. Pickering, Kenneth Kaplan. Prevalence and co-occurrence of substance use disorders and independent mood and anxiety disorders: Results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch. Gen. Psychiatry. 2004;61:807–816. doi: 10.1001/archpsyc.61.8.807. [DOI] [PubMed] [Google Scholar]
- 6.Nehring S.M., Freeman A.M. Alcohol Use Disorder. StatPearls/StatPearls Publishing/StatPearls Publishing LLC.; Treasure Island, FL, USA: 2022. [Google Scholar]
- 7.Hasin D.S., Stinson F.S., Ogburn E., Grant B.F. Prevalence, correlates, disability, and comorbidity of DSM-IV alcohol abuse and dependence in the United States: Results from the National Epidemiologic Survey on Alcohol and Related Conditions. Arch. Gen. Psychiatry. 2007;64:830–842. doi: 10.1001/archpsyc.64.7.830. [DOI] [PubMed] [Google Scholar]
- 8.Rehm J. The risks associated with alcohol use and alcoholism. Alcohol Res. Health. 2011;34:135–143. [PMC free article] [PubMed] [Google Scholar]
- 9.SAMHSA, Center for Behavioral Health Statistics and Quality . 2019 National Survey on Drug Use and Health. Table 2.20B—Binge Alcohol Use in Past Month among Persons Aged 12 or Older, by Age Group and Demographic Characteristics: Percentages, 2018 and 2019. Center for Behavioral Health Statistics and Quality; Rockville, MD, USA: 2019. [Google Scholar]
- 10.Alfonso-Loeches S., Guerri C. Molecular and behavioral aspects of the actions of alcohol on the adult and developing brain. Crit. Rev. Clin. Lab. Sci. 2011;48:19–47. doi: 10.3109/10408363.2011.580567. [DOI] [PubMed] [Google Scholar]
- 11.Sullivan E.V., Desmond J.E., Lim K.O., Pfefferbaum A. Speed and Efficiency but Not Accuracy or Timing Deficits of Limb Movements in Alcoholic Men and Women. Alcohol. Clin. Exp. Res. 2002;26:705–713. doi: 10.1111/j.1530-0277.2002.tb02595.x. [DOI] [PubMed] [Google Scholar]
- 12.Sullivan E.V., Pfefferbaum A. Neurocircuitry in alcoholism: A substrate of disruption and repair. Psychopharmacology. 2005;180:583–594. doi: 10.1007/s00213-005-2267-6. [DOI] [PubMed] [Google Scholar]
- 13.Kane C.J., Drew P.D. Inflammatory responses to alcohol in the CNS: Nuclear receptors as potential therapeutics for alcohol-induced neuropathologies. J. Leukoc. Biol. 2016;100:951–959. doi: 10.1189/jlb.3MR0416-171R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deak T., Kelliher K.T., Wojcik H.J., Gano A. Prenatal and adolescent alcohol exposure programs immunity across the lifespan: CNS-mediated regulation. Pharmacol. Biochem. Behav. 2022;216:173390. doi: 10.1016/j.pbb.2022.173390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alfonso-Loeches S., Pascual-Lucas M., Blanco A.M., Sanchez-Vera I., Guerri C. Pivotal role of TLR4 receptors in alcohol-induced neuroinflammation and brain damage. J. Neurosci. 2010;30:8285–8295. doi: 10.1523/JNEUROSCI.0976-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crews F.T., Qin L., Sheedy D., Vetreno R.P., Zou J. High mobility group box 1/Toll-like receptor danger signaling increases brain neuroimmune activation in alcohol dependence. Biol. Psychiatry. 2013;73:602–612. doi: 10.1016/j.biopsych.2012.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall S.A., McClain J.A., Kelso M.L., Hopkins D.M., Pauly J.R., Nixon K. Microglial activation is not equivalent to neuroinflammation in alcohol-induced neurodegeneration: The importance of microglia phenotype. Neurobiol. Dis. 2013;54:239–251. doi: 10.1016/j.nbd.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin L., He J., Hanes R.N., Pluzarev O., Hong J.S., Crews F.T. Increased systemic and brain cytokine production and neuroinflammation by endotoxin following ethanol treatment. J. Neuroinflamm. 2008;5:10. doi: 10.1186/1742-2094-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kane C.J.M., Phelan K.D., Douglas J.C., Wagoner G., Johnson J.W., Xu J., Phelan P.S., Drew P.D. Effects of Ethanol on Immune Response in the Brain: Region-Specific Changes in Adolescent Versus Adult Mice. Alcohol. Clin. Exp. Res. 2014;38:384–391. doi: 10.1111/acer.12244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ki S.H., Park O., Zheng M., Morales-Ibanez O., Kolls J.K., Bataller R., Gao B. Interleukin-22 treatment ameliorates alcoholic liver injury in a murine model of chronic-binge ethanol feeding: Role of signal transducer and activator of transcription 3. Hepatology. 2010;52:1291–1300. doi: 10.1002/hep.23837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bertola A., Mathews S., Ki S.H., Wang H., Gao B. Mouse model of chronic and binge ethanol feeding (the NIAAA model) Nat. Protoc. 2013;8:627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niedzwiedz-Massey V.M., Douglas J.C., Rafferty T., Johnson J.W., Holloway K.N., Berquist M.D., Kane C.J., Drew P.D. Effects of chronic and binge ethanol administration on mouse cerebellar and hippocampal neuroinflammation. Am. J. Drug Alcohol Abus. 2022:1–14. doi: 10.1080/00952990.2022.2128361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinson M.R., Holloway K.N., Douglas J.C., Kane C.J.M., Miranda R.C., Drew P.D. Divergent and overlapping hippocampal and cerebellar transcriptome responses following developmental ethanol exposure during the secondary neurogenic period. Alcohol. Clin. Exp. Res. 2021;45:1408–1423. doi: 10.1111/acer.14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liao Y., Smyth G.K., Shi W. Feature counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 25.Robinson M.D., Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blighe K., Rana S., Lewis M. EnhancedVolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. 2022.
- 28.Jew B., Alvarez M., Rahmani E., Miao Z., Ko A., Garske K.M., Sul J.H., Pietiläinen K.H., Pajukanta P., Halperin E. Accurate estimation of cell composition in bulk expression through robust integration of single-cell information. Nat. Commun. 2020;11:1971. doi: 10.1038/s41467-020-15816-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeisel A., Muñoz-Manchado A.B., Codeluppi S., Lönnerberg P., La Manno G., Juréus A., Marques S., Munguba H., He L., Betsholtz C., et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 30.Artegiani B., Lyubimova A., Muraro M., van Es J.H., van Oudenaarden A., Clevers H. A Single-Cell RNA Sequencing Study Reveals Cellular and Molecular Dynamics of the Hippocampal Neurogenic Niche. Cell Rep. 2017;21:3271–3284. doi: 10.1016/j.celrep.2017.11.050. [DOI] [PubMed] [Google Scholar]
- 31.Ochocka N., Kaminska B. Microglia Diversity in Healthy and Diseased Brain: Insights from Single-Cell Omics. Int. J. Mol. Sci. 2021;22:3027. doi: 10.3390/ijms22063027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jurga A.M., Paleczna M., Kuter K.Z. Overview of General and Discriminating Markers of Differential Microglia Phenotypes. Front. Cell. Neurosci. 2020;14:198. doi: 10.3389/fncel.2020.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sousa C., Golebiewska A., Poovathingal S.K., Kaoma T., Pires-Afonso Y., Martina S., Coowar D., Azuaje F., Skupin A., Balling R., et al. Single-cell transcriptomics reveals distinct inflammation-induced microglia signatures. EMBO Rep. 2018;19:e46171. doi: 10.15252/embr.201846171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T.K., David E., Baruch K., Lara-Astaiso D., Toth B., et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell. 2017;169:1276–1290.e17. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]
- 35.Krasemann S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z., et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity. 2017;47:566–581.e9. doi: 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsudaira T., Prinz M. Life and death of microglia: Mechanisms governing microglial states and fates. Immunol. Lett. 2022;245:51–60. doi: 10.1016/j.imlet.2022.04.001. [DOI] [PubMed] [Google Scholar]
- 37.Butovsky O., Weiner H.L. Microglial signatures and their role in health and disease. Nat. Rev. Neurosci. 2018;19:622–635. doi: 10.1038/s41583-018-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Das S., Li Z., Noori A., Hyman B.T., Serrano-Pozo A. Meta-analysis of mouse transcriptomic studies supports a context-dependent astrocyte reaction in acute CNS injury versus neurodegeneration. J. Neuroinflamm. 2020;17:227. doi: 10.1186/s12974-020-01898-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L.E., La Manno G., et al. Molecular Architecture of the Mouse Nervous System. Cell. 2018;174:999–1014.e22. doi: 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McClain J.A., Morris S.A., Deeny M.A., Marshall S.A., Hayes D.M., Kiser Z.M., Nixon K. Adolescent binge alcohol exposure induces long-lasting partial activation of microglia. Brain Behav. Immun. 2011;25:S120–S128. doi: 10.1016/j.bbi.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He J., Crews F.T. Increased MCP-1 and microglia in various regions of the human alcoholic brain. Exp. Neurol. 2008;210:349–358. doi: 10.1016/j.expneurol.2007.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sofroniew M.V., Vinters H.V. Astrocytes: Biology and pathology. Acta Neuropathol. 2010;119:7–35. doi: 10.1007/s00401-009-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verkhratsky A., Nedergaard M. Physiology of Astroglia. Physiol. Rev. 2018;98:239–389. doi: 10.1152/physrev.00042.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Magistretti P.J., Allaman I. Lactate in the brain: From metabolic end-product to signalling molecule. Nat. Rev. Neurosci. 2018;19:235–249. doi: 10.1038/nrn.2018.19. [DOI] [PubMed] [Google Scholar]
- 45.Nicola J., Allen C.E. Cell Biology of Astrocyte-Synapse Interactions. Neuron. 2017;96:697–708. doi: 10.1016/j.neuron.2017.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khakh B.S. Astrocyte–Neuron Interactions in the Striatum: Insights on Identity, Form, and Function. Trends Neurosci. 2019;42:617–630. doi: 10.1016/j.tins.2019.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dong Y., Benveniste E.N. Immune function of astrocytes. Glia. 2001;36:180–190. doi: 10.1002/glia.1107. [DOI] [PubMed] [Google Scholar]
- 48.Sofroniew M.V. Astrocyte barriers to neurotoxic inflammation. Nat. Rev. Neurosci. 2015;16:249–263. doi: 10.1038/nrn3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vallés S.L., Blanco A.M., Pascual M., Guerri C. Chronic ethanol treatment enhances inflammatory mediators and cell death in the brain and in astrocytes. Brain Pathol. 2004;14:365–371. doi: 10.1111/j.1750-3639.2004.tb00079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Blanco A.M., Pascual M., Valles S.L., Guerri C. Ethanol-induced iNOS and COX-2 expression in cultured astrocytes via NF-κB. NeuroReport. 2004;15:681–685. doi: 10.1097/00001756-200403220-00021. [DOI] [PubMed] [Google Scholar]
- 51.Alfonso-Loeches S., Pascual M., Gómez-Pinedo U., Pascual-Lucas M., Renau-Piqueras J., Guerri C. Toll-like receptor 4 participates in the myelin disruptions associated with chronic alcohol abuse. Glia. 2012;60:948–964. doi: 10.1002/glia.22327. [DOI] [PubMed] [Google Scholar]
- 52.Vargas W.M., Bengston L., Gilpin N.W., Whitcomb B.W., Richardson H.N. Alcohol Binge Drinking during Adolescence or Dependence during Adulthood Reduces Prefrontal Myelin in Male Rats. J. Neurosci. 2014;34:14777–14782. doi: 10.1523/JNEUROSCI.3189-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zahr N.M., Luong R., Sullivan E.V., Pfefferbaum A. Measurement of serum, liver, and brain cytokine induction, thiamine levels, and hepatopathology in rats exposed to a 4-day alcohol binge protocol. Alcohol. Clin. Exp. Res. 2010;34:1858–1870. doi: 10.1111/j.1530-0277.2010.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pascual M., Baliño P., Alfonso-Loeches S., Aragón C.M., Guerri C. Impact of TLR4 on behavioral and cognitive dysfunctions associated with alcohol-induced neuroinflammatory damage. Brain Behav Immun. 2011;25((Suppl. 1)):S80–S91. doi: 10.1016/j.bbi.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 55.Pascual M., Baliño P., Aragón C.M., Guerri C. Cytokines and chemokines as biomarkers of ethanol-induced neuroinflammation and anxiety-related behavior: Role of TLR4 and TLR2. Neuropharmacology. 2015;89:352–359. doi: 10.1016/j.neuropharm.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 56.Ruffle J.K. Molecular neurobiology of addiction: What’s all the (Δ)FosB about? Am. J. Drug Alcohol Abus. 2014;40:428–437. doi: 10.3109/00952990.2014.933840. [DOI] [PubMed] [Google Scholar]
- 57.Wille-Bille A., de Olmos S., Marengo L., Chiner F., Pautassi R.M. Long-term ethanol self-administration induces ΔFosB in male and female adolescent, but not in adult, Wistar rats. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2017;74:15–30. doi: 10.1016/j.pnpbp.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 58.Wille-Bille A., Marengo L., Godino A., Pautassi R.M. Effects of escalating versus fixed ethanol exposure on ∆FosB expression in the mesocorticolimbic pathway in adolescent and adult rats. Am. J. Drug Alcohol Abus. 2021;47:569–580. doi: 10.1080/00952990.2021.1954188. [DOI] [PubMed] [Google Scholar]
- 59.Montesinos J., Pascual M., Rodríguez-Arias M., Miñarro J., Guerri C. Involvement of TLR4 in the long-term epigenetic changes, rewarding and anxiety effects induced by intermittent ethanol treatment in adolescence. Brain Behav. Immun. 2016;53:159–171. doi: 10.1016/j.bbi.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 60.Nomaru H., Sakumi K., Katogi A., Ohnishi Y.N., Kajitani K., Tsuchimoto D., Nestler E.J., Nakabeppu Y. Fosb gene products contribute to excitotoxic microglial activation by regulating the expression of complement C5a receptors in microglia. Glia. 2014;62:1284–1298. doi: 10.1002/glia.22680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shen H., French B.A., Liu H., Tillman B.C., French S.W. Increased activity of the complement system in the liver of patients with alcoholic hepatitis. Exp. Mol. Pathol. 2014;97:338–344. doi: 10.1016/j.yexmp.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCullough R.L., McMullen M.R., Das D., Roychowdhury S., Strainic M.G., Medof M.E., Nagy L.E. Differential contribution of complement receptor C5aR in myeloid and non-myeloid cells in chronic ethanol-induced liver injury in mice. Mol. Immunol. 2016;75:122–132. doi: 10.1016/j.molimm.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCullough R.L., McMullen M.R., Poulsen K.L., Kim A., Medof M.E., Nagy L.E. Anaphylatoxin Receptors C3aR and C5aR1 Are Important Factors That Influence the Impact of Ethanol on the Adipose Secretome. Front. Immunol. 2018;9:2133. doi: 10.3389/fimmu.2018.02133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kalinin S., González-Prieto M., Scheiblich H., Lisi L., Kusumo H., Heneka M.T., Madrigal J.L.M., Pandey S.C., Feinstein D.L. Transcriptome analysis of alcohol-treated microglia reveals downregulation of beta amyloid phagocytosis. J. Neuroinflamm. 2018;15:141. doi: 10.1186/s12974-018-1184-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kane C.J.M., Douglas J.C., Rafferty T., Johnson J.W., Niedzwiedz-Massey V.M., Phelan K.D., Majewska A.K., Drew P.D. Ethanol modulation of cerebellar neuroinflammation in a postnatal mouse model of fetal alcohol spectrum disorders. J. Neurosci. Res. 2021;99:1986–2007. doi: 10.1002/jnr.24797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drew P.D., Johnson J.W., Douglas J.C., Phelan K.D., Kane C.J.M. Pioglitazone Blocks Ethanol Induction of Microglial Activation and Immune Responses in the Hippocampus, Cerebellum, and Cerebral Cortex in a Mouse Model of Fetal Alcohol Spectrum Disorders. Alcohol. Clin. Exp. Res. 2015;39:445–454. doi: 10.1111/acer.12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang Y., Le W. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Mol. Neurobiol. 2016;53:1181–1194. doi: 10.1007/s12035-014-9070-5. [DOI] [PubMed] [Google Scholar]
- 68.Franco R., Fernández-Suárez D. Alternatively activated microglia and macrophages in the central nervous system. Prog. Neurobiol. 2015;131:65–86. doi: 10.1016/j.pneurobio.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 69.Paolicelli R.C., Sierra A., Stevens B., Tremblay M.-E., Aguzzi A., Ajami B., Amit I., Audinat E., Bechmann I., Bennett M., et al. Microglia states and nomenclature: A field at its crossroads. Neuron. 2022;110:3458–3483. doi: 10.1016/j.neuron.2022.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Moore C.S., Ase A.R., Kinsara A., Rao V.T., Michell-Robinson M., Leong S.Y., Butovsky O., Ludwin S.K., Séguéla P., Bar-Or A., et al. P2Y12 expression and function in alternatively activated human microglia. Neurol.-Neuroimmunol. Neuroinflamm. 2015;2:e80. doi: 10.1212/NXI.0000000000000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Holtman I.R., Raj D.D., Miller J.A., Schaafsma W., Yin Z., Brouwer N., Wes P.D., Möller T., Orre M., Kamphuis W., et al. Induction of a common microglia gene expression signature by aging and neurodegenerative conditions: A co-expression meta-analysis. Acta Neuropathol. Commun. 2015;3:31. doi: 10.1186/s40478-015-0203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Naj A.C., Jun G., Reitz C., Kunkle B.W., Perry W., Park Y.S., Beecham G.W., Rajbhandary R.A., Hamilton-Nelson K.L., Wang L.-S., et al. Effects of multiple genetic loci on age at onset in late-onset Alzheimer disease: A genome-wide association study. JAMA Neurol. 2014;71:1394–1404. doi: 10.1001/jamaneurol.2014.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hickman S.E., Kingery N.D., Ohsumi T.K., Borowsky M.L., Wang L.-C., Means T.K., El Khoury J. The microglial sensome revealed by direct RNA sequencing. Nat. Neurosci. 2013;16:1896–1905. doi: 10.1038/nn.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ransohoff R.M., Brown M.A. Innate immunity in the central nervous system. J. Clin. Investig. 2012;122:1164–1171. doi: 10.1172/JCI58644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zamanian J.L., Xu L., Foo L.C., Nouri N., Zhou L., Giffard R.G., Barres B.A. Genomic Analysis of Reactive Astrogliosis. J. Neurosci. 2012;32:6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liddelow S.A., Guttenplan K.A., Clarke L.E., Bennett F.C., Bohlen C.J., Schirmer L., Bennett M.L., Münch A.E., Chung W.-S., Peterson T.C., et al. Neurotoxic reactive astrocytes are induced by activated microglia. Nature. 2017;541:481–487. doi: 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Blanco A.M., Vallés S.L., Pascual M., Guerri C. Involvement of TLR4/type I IL-1 receptor signaling in the induction of inflammatory mediators and cell death induced by ethanol in cultured astrocytes. J. Immunol. 2005;175:6893–6899. doi: 10.4049/jimmunol.175.10.6893. [DOI] [PubMed] [Google Scholar]
- 78.Floreani N.A., Rump T.J., Muneer P.M.A., Alikunju S., Morsey B.M., Brodie M.R., Persidsky Y., Haorah J. Alcohol-induced interactive phosphorylation of Src and toll-like receptor regulates the secretion of inflammatory mediators by human astrocytes. J. Neuroimmune Pharmacol. 2010;5:533–545. doi: 10.1007/s11481-010-9213-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alfonso-Loeches S., Ureña-Peralta J.R., Morillo-Bargues M.J., Oliver-De La Cruz J., Guerri C. Role of mitochondria ROS generation in ethanol-induced NLRP3 inflammasome activation and cell death in astroglial cells. Front. Cell. Neurosci. 2014;8:216. doi: 10.3389/fncel.2014.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baumann N., Pham-Dinh D. Biology of Oligodendrocyte and Myelin in the Mammalian Central Nervous System. Physiol. Rev. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- 81.El Waly B., Macchi M., Cayre M., Durbec P. Oligodendrogenesis in the normal and pathological central nervous system. Front. Neurosci. 2014;8:145. doi: 10.3389/fnins.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wozniak J.R., Riley E.P., Charness M.E. Clinical presentation, diagnosis, and management of fetal alcohol spectrum disorder. Lancet Neurol. 2019;18:760–770. doi: 10.1016/S1474-4422(19)30150-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harper C. The neuropathology of alcohol-related brain damage. Alcohol Alcohol. 2009;44:136–140. doi: 10.1093/alcalc/agn102. [DOI] [PubMed] [Google Scholar]
- 84.Monnig M.A., Yeo R.A., Tonigan J.S., McCrady B.S., Thoma R.J., Sabbineni A., Hutchison K.E. Associations of White Matter Microstructure with Clinical and Demographic Characteristics in Heavy Drinkers. PLoS ONE. 2015;10:e0142042. doi: 10.1371/journal.pone.0142042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Back S.A., Gan X., Li Y., Rosenberg P.A., Volpe J.J. Maturation-Dependent Vulnerability of Oligodendrocytes to Oxidative Stress-Induced Death Caused by Glutathione Depletion. J. Neurosci. 1998;18:6241–6253. doi: 10.1523/JNEUROSCI.18-16-06241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rice J., Gu C. Function and Mechanism of Myelin Regulation in Alcohol Abuse and Alcoholism. BioEssays. 2019;41:1800255. doi: 10.1002/bies.201800255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Luo J., Miller M.W. Growth factor-mediated neural proliferation: Target of ethanol toxicity. Brain Res. Rev. 1998;27:157–167. doi: 10.1016/S0165-0173(98)00009-5. [DOI] [PubMed] [Google Scholar]
- 88.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus [88] and are accessible through GEO Series accession number GSE222445 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE222445, accessed on 24 February 2023).