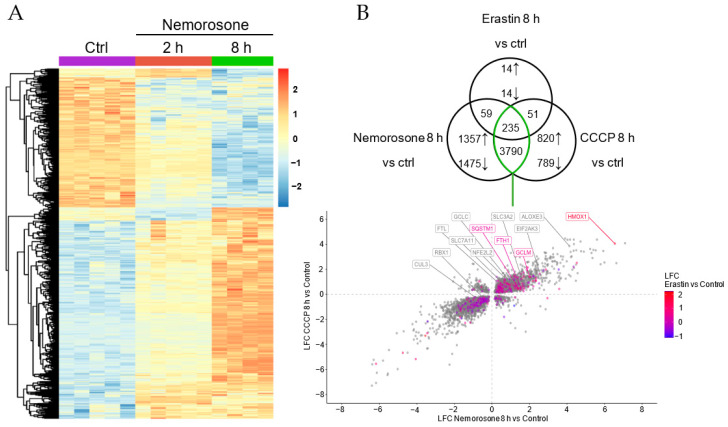
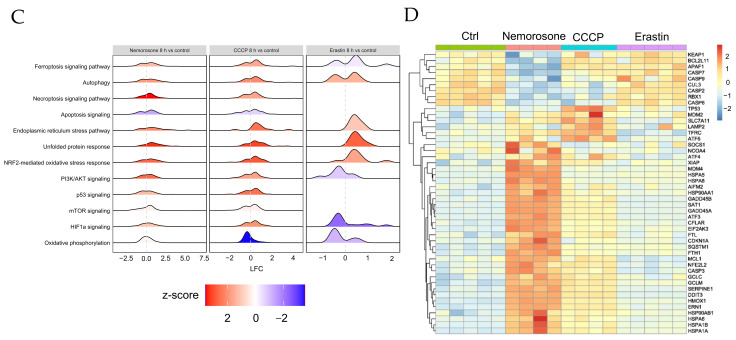
Figure 4.
Genes and signaling pathways differentially expressed after treatment with nemorosone, CCCP, and erastin in HT1080 cells. (A) Heatmap of the expression of genes that are differentially expressed after both nemorosone (100 μM) 2 h and nemorosone (100 μM) 8 h treatments, compared to the control. Each column represents a sample; each row represents a gene. The genes have been clustered, and their expression has been centered and scaled. (B) Venn diagram showing differentially expressed (DE) genes, compared to the control, regulated by one, two, or all three treatments at the same time point (nemorosone 100 μM, CCCP 50 μM, erastin 20 μM; 8 h). The direction of the arrows indicates upregulation or downregulation in the case of individual treatments. Each intersection shows the set of genes modulated by two or three treatments. The scatterplot below was made by plotting the LFC values (log2fold change), metric indicating how much the expression of a gene changes compared to the control condition of the corresponding genes indicated by the green intersection: positive values indicate higher expression than the control, and negative values indicate lower expression than the control. DE genes by erastin are colored according to their LFC value. Known ferroptosis-related genes are labeled. (C) Ridge plots showing the IPA (Ingenuity Pathway Analysis) pathways that were found to be significantly enriched in DE gene sets. Each density plot shows the distribution of the LFC values of the DE genes involved in the enriched pathway. Density plots are scaled per panel. The color indicates the z-score, which is an IPA metric that predicts activation (positive values) or inhibition (negative values) of the pathway. (D) Heatmap of the expression of selected ferroptosis-related genes. Each column represents a sample; each row represents a gene. The genes have been clustered, and their expression has been centered and scaled.