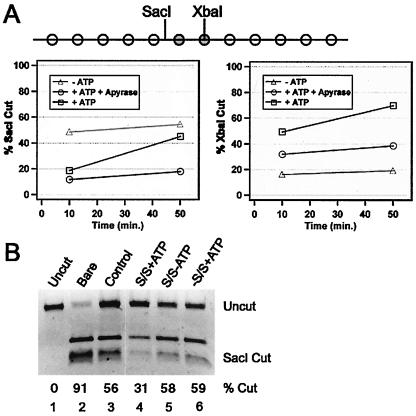
FIG. 3.
5S nucleosomal arrays are stably remodeled by hSWI/SNF. (A) Diagram of the 5S-G5E4 rDNA array used. White circles represent nucleosomes at preferred sites on 208-bp 5S rDNA sequences. Grey circles represent nucleosomes in the transcription template. Locations of SacI and XbaI sites relative to inferred unremodeled nucleosome positions are indicated. For the graphs, the array was treated with hSWI/SNF without ATP (triangles), with ATP for 30 min (squares), or with ATP for 30 min, followed by apyrase for 18 min (circles), and then cut with SacI (left) or XbaI (right) for the indicated times. The purified DNA was separated by agarose electrophoresis, and percent cutting was quantified. Similar results were obtained in three separate experiments. (B) Unlabeled arrays were incubated with both SWI/SNF (S/S) and ATP (lane 4), without ATP (lane 5), or without SWI/SNF (lane 6) for 90 min before dialysis into TE. Samples of these reaction mixtures and bare DNA (lane 2) or the untreated assembled array (lane 3) were digested with SacI, purified, and separated as described above, followed by ethidium bromide staining and quantitation. Lane 1 contained uncut DNA.