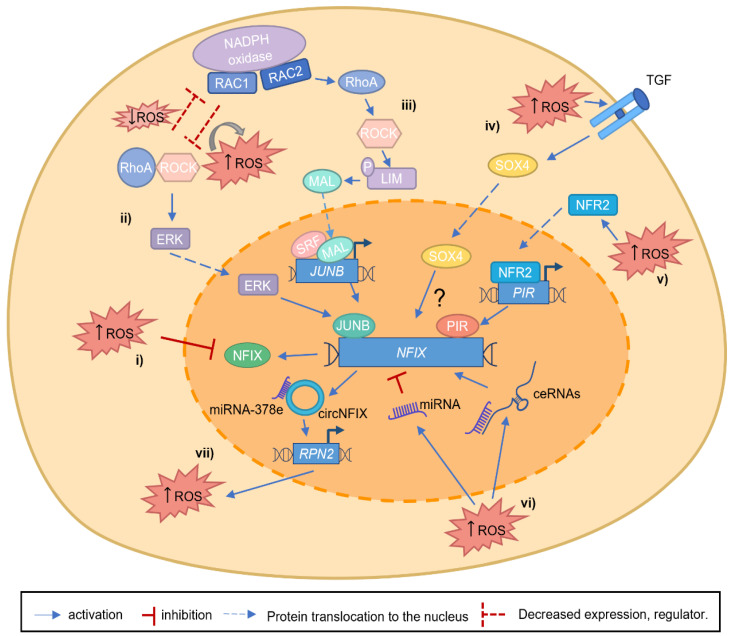
Figure 3.
Putative mechanisms linking NFIX and increased ROS levels. (i) During tissue development, ROS can regulate NFIX expression, oxidizing specific NFIX cysteine residues. (ii). RAC1, a catalytic subunit of NADPH oxidase, can generate ROS and interact with RhoA/ROCK axis leading to its activation or inhibition. On the other hand, RhoA, through its target gene ROCK, can inhibit RAC1 activity, decreasing ROS levels. When RAC1 is able to activate the RhoA/ROCK axis and increase ROS levels, ERK kinases are activated, leading to JUNB activation, and consequently, JUNB activates NFIX expression. (iii) The increased expression of RAC2, and subsequent ROS production, lead to the activation and increase in RhoA and its targets, such as ROCK, that will phosphorylate LIM kinase. With LIM kinase phosphorylation, MAL translocates to the nucleus activating SRF. Consequently, SRF activates JUNB expression, and JUNB protein binds to the NFIX promoter, activating its transcription. (iv) NFIX is a putative target gene of SOX4, but the exact mechanism remains unknown (question mark). SOX4 activation is mediated by ROS/TGFβ, and it is possible that this protein can translocate to the nucleus activating NFIX expression. (v). The oxidative stress sensor NRF2 is activated by increased ROS levels, allowing its translocation to the nucleus, where it activates the Pirin (PIR) promoter. Consequently, NFIX expression is induced through the binding of Pirin to the NFI I/CCAAT box transcription factor (NFI/CTF1) domain. (vi) ROS can lead to the activation of miRNAs or ceRNAs (network composed by miRNA and the inhibitory binding through lncRNA), which lead to a decrease or an increase in NFIX expression, respectively. (vii) In gliomas, miR-378e functionally targets RPN2, an oncogene, and ROS inducer, inhibiting its oncogenic functions. circNFIX can sponge the action of miR-378e, allowing RPN2 activity (as part of the ceRNA network), contributing to glioma progression.