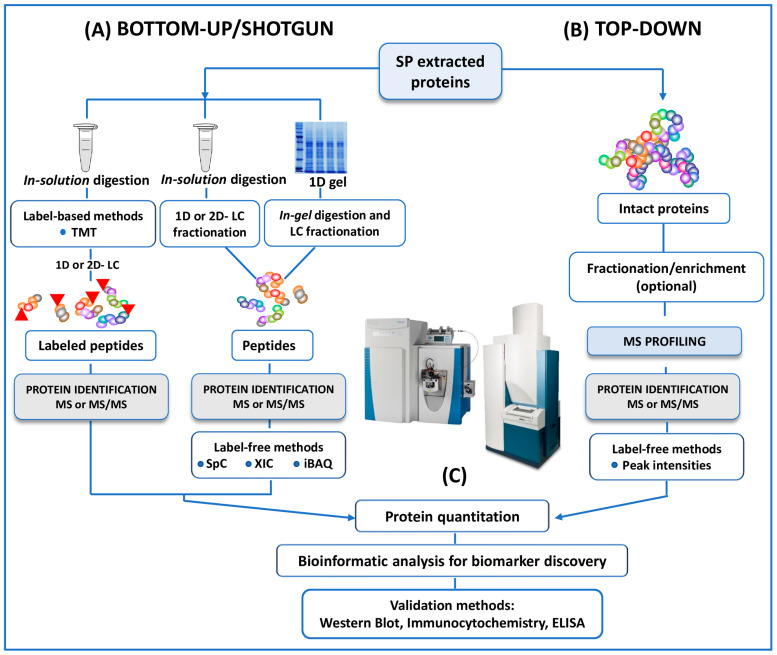
Figure 2.
A schematic outline of all the protocols here reviewed in both bottom-up and top-down untargeted MS approaches for the proteomic analysis of human SP. (A) In conventional bottom-up (or shotgun) strategy, proteins extracted from SP are reduced, alkylated and digested in solution. The resulting proteolytic fragments are fractionated by 1D- or 2D- LC before MS analysis. In a one more classical approach, included in the category of the bottom-up strategies, SP proteins are separated by 1D (here reviewed) or 2D SDS-PAGE then the bands (from 1D) or the spots (from 2D) of interest are enzymatically cleaved by the mean of “in gel digestion” protocols. Protein quantitation, when requested, could be performed by labeling strategies (iTRAQ or other TMT strategies). In this case, proteolytic peptides are labeled with isobaric mass tags and pooled together prior to HPLC separation. Multiplexed quantitation of changes in SP protein expression is measured by MS and MS/MS experiments. MS-quantitation in several studies is assessed alternatively by label-free methods (SpC, XIC or iBAQ). (B) In the top-down investigations here reviewed no digestion experiments are performed. The mixture of intact proteins from SP is fractionated, or enriched and/ or fractionated before MS or MS/MS analysis. The quantification is performed by label-free methods in the studies. (C) Identified (and or quantified) proteins are analyzed by bioinformatics tools for the extrapolation of differentially expressed proteins as candidate biomarkers of male infertility. Putative biomarkers from MS analysis are then validated using different approaches, including Western Blot, immunocytochemistry, ELISA, Selected reaction monitoring/Multiple Reaction Monitoring.