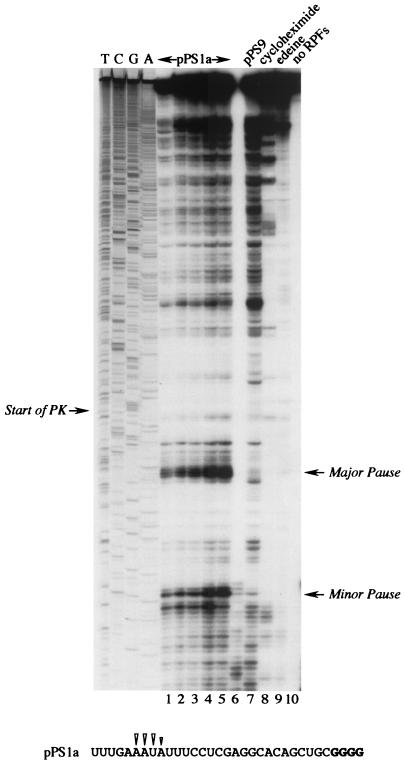
FIG. 2.
Ribosomal pausing at the minimal IBV pseudoknot. mRNA from AvaII-digested pPS1a was subjected to heelprint analysis as detailed in Materials and Methods. Heelprints of the minimal IBV pseudoknot (pPS1a; lanes 1 to 6 and 8 to 10) and a mutant derivative (pPS9, lane 7) are shown alongside a sequencing ladder (TCGA). Each reaction mixture contained 20 ng of the relevant single-stranded DNA template. In lanes 1 to 5, the concentrations of primer and RPFs were varied: lane 1, 0.1 ng of primer, 3 μl of RPFs; lane 2, 0.2 ng of primer, 3 μl of RPFs; lane 3, 0.4 ng of primer, 3 μl of RPFs; lane 4, 0.4 ng of primer, 4 μl of RPF; lane 5, 0.4 ng of primer, 4.5 μl of RPFs. All other lanes (except lane 10) contained 0.4 ng of primer and 3 μl of RPFs. Lanes 8 and 9 were control reactions in which cycloheximide (lane 8) or edeine (lane 9) was added (to 1 mM and 5 μM concentrations, respectively) prior to addition of mRNA to the translation reaction mixture. In lane 10, RPFs were omitted from the primer extension reaction. The start of the pseudoknot (the first G in a block of four reading up the gel) and the position of two clear pause sites are indicated with arrows. Lane 6 was identical to lane 5, except T4 DNA polymerase replaced T7 DNA polymerase (unsuccessfully). The primary sequence of the mRNA upstream of the pseudoknot is shown at the bottom, and the positions of the pseudoknot-dependent heelprints are indicated with arrowheads (the first four G residues of the pseudoknot are shown in bold type).