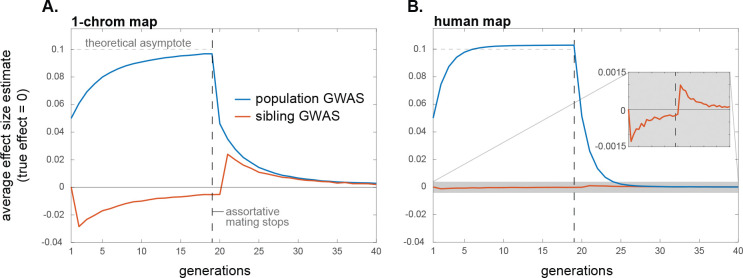
Figure 2:
Assortative mating systematically biases effect size estimation in population and within-family GWASs, although the bias in within-family GWASs is expected usually to be small. Here, cross-trait assortative mating between traits 1 and 2 occurs for the first 19 generations, after which mating is random. Assortative mating is sex-asymmetric, with strength . Distinct sets of loci underlie variation in trait 1 and 2, with effect sizes at causal loci normalized to 1. Plotted are average estimated effects of the focal alleles at loci causal for trait 1 in population and within-family GWASs on trait 2, for a hypothetical genome with one chromosome of length 1 Morgan (A) and for humans (B). Since the traits have distinct genetic bases, the true effects on trait 2 of the alleles at trait-1 loci are zero. The horizontal lines at 0.1 are a theoretical ‘first-order’ approximation of the asymptotic bias in a population GWAS (Appendix A3.1). Profiles are averages across 10,000 replicate simulation trials. Simulation details can be found in the Methods.