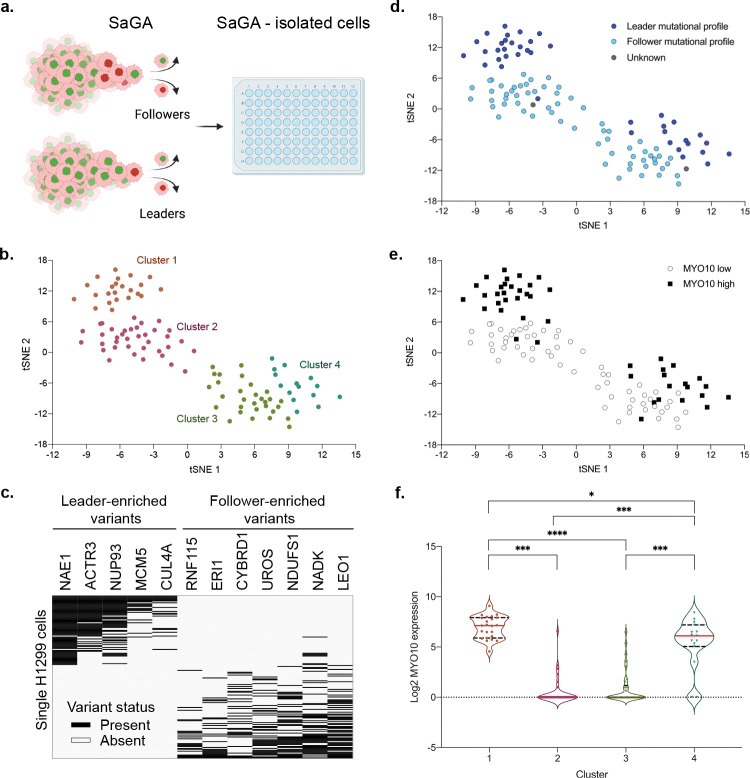
Fig. 6 |. SaGA-isolated single cell RNA sequencing and analysis.
a. Workflow schematic of collection of SaGA-isolated cells for single cell RNA sequencing and analysis. H1299 cells were used to perform a 3D spheroid invasion assay for a 24 h invasion period. Upon application of SaGA, user-defined cell populations (Leaders and Followers) were sorted directly into a 96-well plate with RNA lysis buffer, frozen at − 80 °C, and subsequently processed for single-cell RNA sequencing. Created with Biorender.com. b. tSNE plot of single H1299 cells (n = 105) based upon expression of the most variably expressed genes (n = 1,155). tSNE clusters were determined by cell positioning within the plot. c. Mutation profile plot for n = 171 single cells using previously identified H1299 leader- and follower- specific mutations30. d. tSNE plot with each cell labeled by its mutation profile from panel c. e. tSNE plot with each cell labeled by high (defined as [log2 (normalized counts + 1)] >2 or low <2 expression of Myo10. f. Quantification of Myo10 expression for each tSNE cluster. *p < 0.05 by one-way ANOVA with Tukey’s multiple comparisons test.