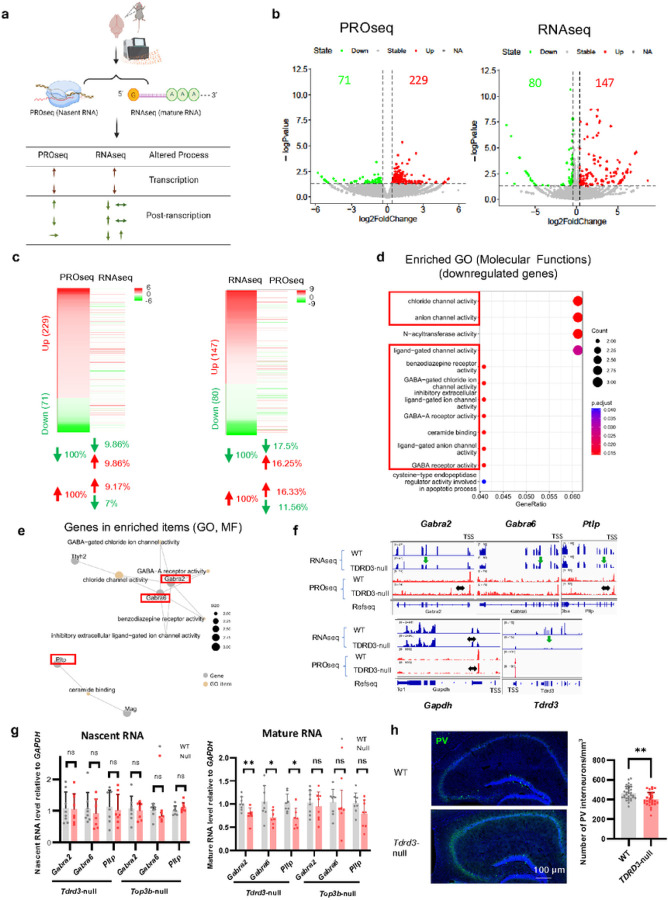
Figure 5. Several mRNAs associated with GABAergic interneurons are downregulated by post-transcriptional mechanisms in Tdrd3-null mice.
a A schematic diagram of experimental design depicts that nascent and mature mRNAs are pro led by PROseq and RNAseq, respectively. The former method detects mRNA altered at the transcriptional step, whereas the latter measures mRNA changes caused by both transcriptional and post-transcriptional mechanisms. The up or down arrows indicate differentially expressed genes (DEGs) that are increased or decreased in null mice, respectively, whereas the horizonal arrows mark mRNAs that remain unchanged. The mRNAs that do not show the same direction of alteration in both methods should be regulated by post-transcriptional mechanism (such as turnover). b Volcano plots show differentially expressed genes (DEGs) identified by PROseq (left) and RNAseq (right) in Tdrd3 null mouse brains (green, downregulated; red, upregulated, log2FoldChange > 1.3 and p-value < 0.05). c HEATmap analysis to compare DEGs obtained by PROseq vs. those by RNAseq. The left panel illustrates how the up or down-regulated DEGs from PROseq are altered in RNAseq. The right panel shows the reciprocal comparison: how the up and down-regulated DEGs from RNAseq are altered in PROseq. The data lines marked in the same color indicate DEGs that are altered in the same direction by both assays. The table below the HEATmaps indicate that the percentages of DEGs that are altered in the same or opposite directions, as marked by the directions of arrows. d Gene ontology analysis of downregulated DEGs from RNAseq using molecular function category indicates that the expression of genes associated with chloride channels in GABAergic interneurons (marked by red boxes) is disturbed in Tdrd3-nullmice. e Relationship between several enriched GO terms and their associated genes from (d). f Graphs from USCS genome browser show that several representative genes associated with chloride channels exhibit reduced signals by RNAseq but unchanged signals by PROseq (Gabra2, Gabra6, Pltp) in Tdrd3-null mice. The down arrows mark the RNAseq signals thar are reduced in the null mice, whereas the horizontal arrows mark the PROseq signals that are unchanged. Gapdh and Tdrd3 genes are included as negative and positive controls, respectively. g RT-qPCR results of nascent RNA (left) and mature RNA (right) expression of Gabra2, Gabra6 and Pltp in brains of Tdrd3-null mice and Top3b-null mice. h Immunofluorescent images (left) and their Quantification (right) show that the density of parvalbumin positive cells is significantly reduced in Tdrd3-nullmice. Data are presented as mean values + SD. Two-tailed Student’s t test was performed for the comparisons. p-values < 0.05, 0.01 are marked as: *, **; p-value > 0.5 is marked as ns.