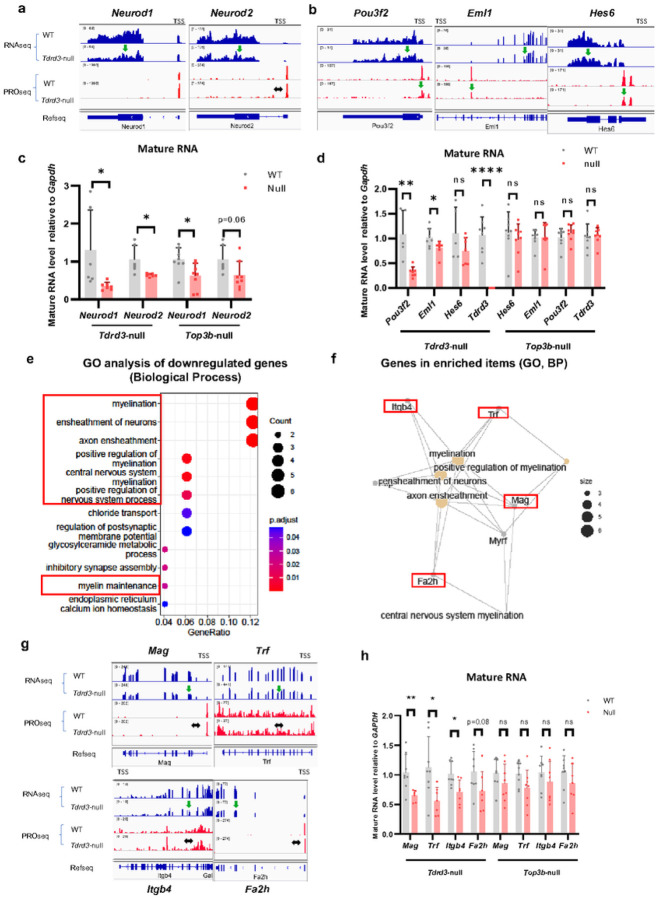
Figure 6. Several Gabra2 downstream genes and myelination associated genes are downregulated by post-transcriptional mechanism in Tdrd3-nullmice.
a Bedgraphs from UCSC genome browser analysis show that two genes downstream of Gabra2, Neurod1 and Neurod2, exhibit reduced RNAseq signals but unchanged PROseq signals in Tdrd3-null mice. b Bedgraphs from UCSC genome browser analysis show that three genes that are known to be bound and enhanced expression by Neurod1 exhibit reduced signals of both RNAseq and PROseq in Tdrd3-nullmice. The down arrows mark reduced signals, whereas horizontal arrows mark genes that are unchanged in the null mice. c RT-qPCR results show that mature RNA levels of Neurod1 and Neurod2 are significantly decreased in Tdrd3-null mice (p<0.05), and both genes also show a strong trend of reduction in Top3b-null mice (for Neurod2, p=0.06; which does not reach statistical significance). d RT-qPCR analyses show that mature RNA levels of Eml1 and Pou3f2, but not Hes6, are significantly reduced in Tdrd3-nullmice. e. Gene ontology enrichment analysis of downregulated genes in Tdrd3-null mice using Biological Process category shows that several top enriched GO terms are related to myelination (marked in red boxes). f Relationship between enriched GO items and their associated genes reveals that these terms are closely associated with each other and share several common genes (marked as red rectangles). g Bedgraphs from USSC genome browser analysis of RNAseq and PROseq show that mature but not nascent RNA levels for several myelination-associated genes are reduced in Tdrd3-null mice (Mag, Trf, Itgb4 and Fa2h). The down arrows mark reduced signals, whereas horizontal arrows mark genes that are unchanged in the null mice. h RT-qPCR results show that mature RNA levels of several myelination associated genes are decreased in Tdrd3-null, but not Top3b-null mice. Data are presented as mean values + SD. Two-tailed Student’s t test was performed for the comparisons. p-values < 0.05, 0.01, 0.001, and 0.0001 are marked as: *, **, ***, and ****; p-value > 0.5 is marked as ns.