Figure 1: Different patterns of chromatin opening by PU.1 and C/EBP.
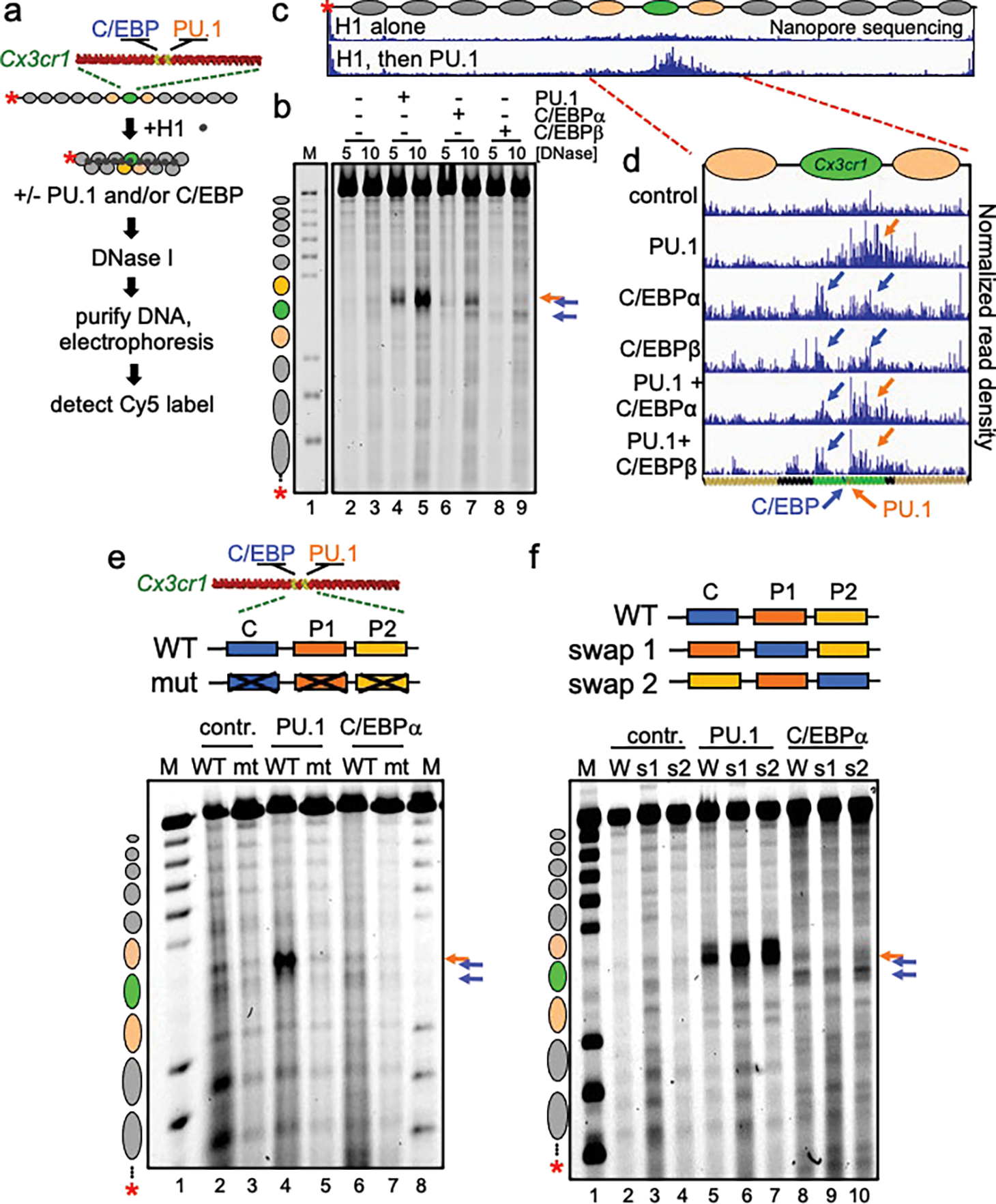
a, Schematic of chromatin assembly, compaction, transcription factor binding, and DNase I digestion assays. b, DNase I digestion analysis of TFs binding to H1-compacted nucleosome arrays visualized by gel electrophoresis at two DNase I concentrations (in ng/uL) Distinct hypersensitive sites are observed, relative to no TF control (lanes 2–3), upon PU.1 (lanes 4–5, orange arrow), C/EBPα (lanes 6–7, blue arrows), and C/EBPβ (lanes 8–9, blue arrows). Lane M, partial EcoRI digest of end-labeled array fragment. Positions of nucleosomes on reconstituted arrays are indicated. c and d, Nanopore sequencing endpoint analysis of DNase I digested (10 ng/uL) H1-compacted arrays. Ovals indicate the translational positions of nucleosomes determined in Extended Data Fig. 2d. Plots show normalized read density on the y axis. For each plot, the maximum value is set to 0.4% of reads. e, DNase I digestion (10 ng/uL) of H1-compacted wild-type Cx3cr1 nucleosome arrays (WT) or motif mutant Cx3cr1 nucleosome arrays (mt) incubated alone (contr., lanes 2–3), with PU.1 (lanes 4–5) or C/EBPα (lanes 6–7). f, DNase I digestion of H1-compacted wild-type Cx3cr1 nucleosome arrays (WT) or PU.1 and C/EBPα motif swap arrays (swap 1 or swap 2) incubated alone (contr., lanes 2–4), with PU.1 (lanes 5–7) or C/EBPα (lanes 8–10).
