Extended Data Figure 8: The IDR domain of PU.1 is most crucial for chromatin opening.
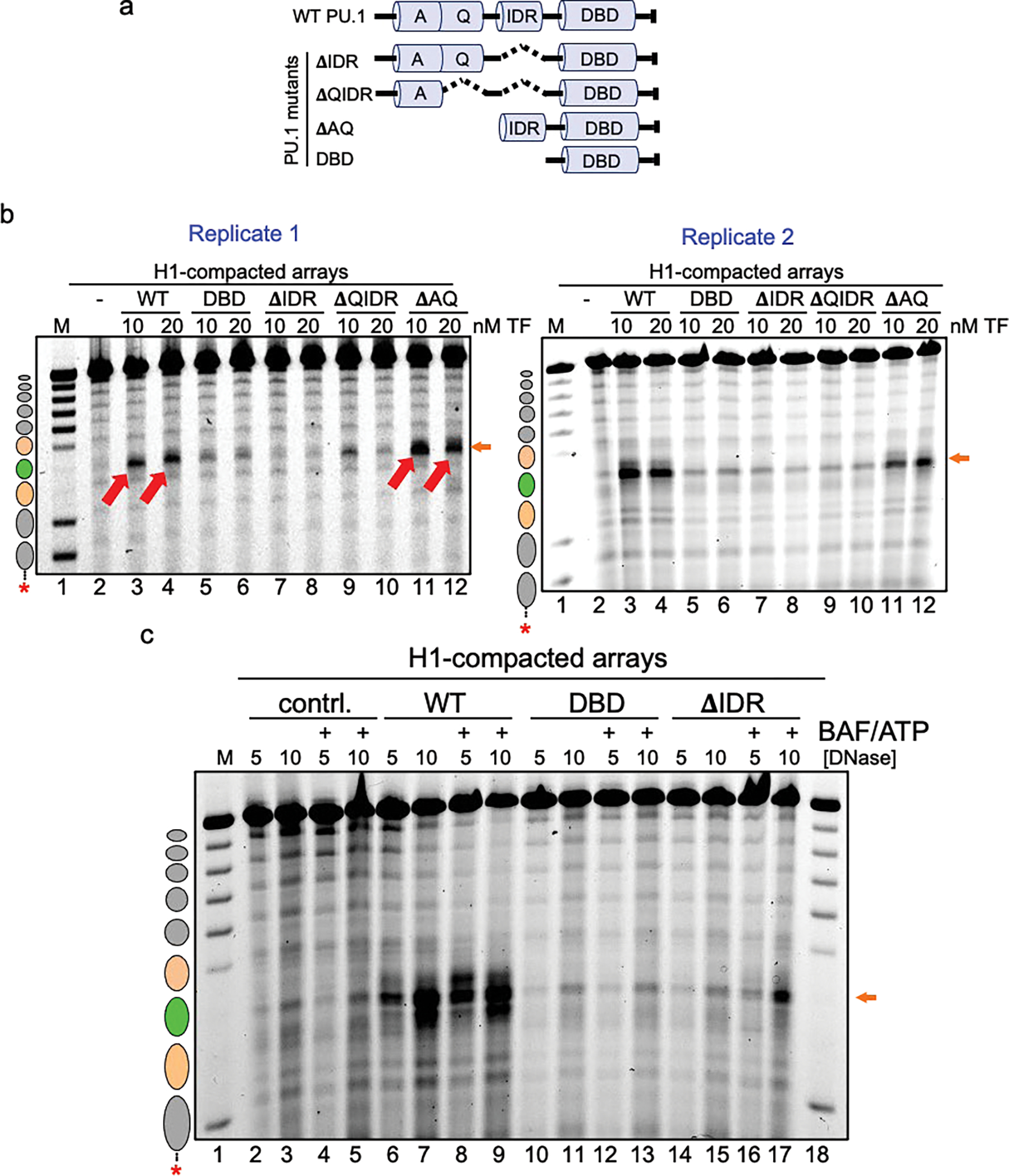
a, Illustration of the PU.1 deletion mutant series. Shown are the positions of the acidic domain (A), Q-rich domain (Q), intrinsically disordered region (IDR) and DNA-binding domain (DBD). b, DNase I digestion analysis of two transcription factor concentrations binding to H1-compacted nucleosome arrays visualized by gel electrophoresis (10 ng/uL DNase I). c, DNase I digestion analysis of H1-compacted nucleosome arrays incubated with no TFs (contrl., lanes 2–5), WT PU.1 (lanes 6–9), DBD (lanes 10–13), and ΔIDR deletion (lanes 14–17) with or without the addition of cBAF and ATP.
