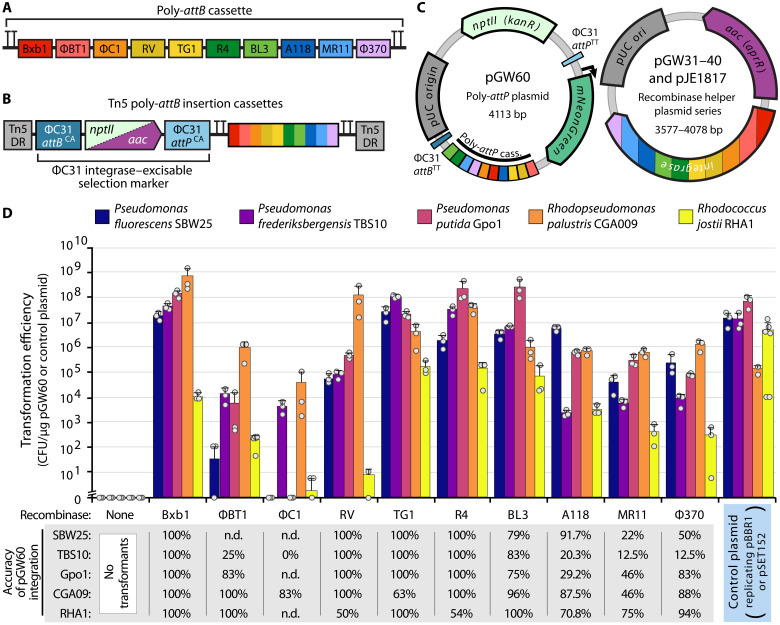
Fig. 2. SAGE enables stable, highly efficient integration of plasmid DNA into the genomes of engineered bacteria.
(A) Diagram of genome-integrated 10× poly-attB cassette, including terminators for transcriptional insulation. Each attB sequence is indicated by a color-coded box and is flanked by a random 20-nt DNA spacer sequence. (B) Diagram of Tn5 poly-attB insertion cassettes. Cassettes are flanked by Tn5 direct repeat (DR) sequences and contain an antibiotic resistance cassette (either nptII or aac) upstream of the poly-attB cassette from (A). A cognate pair of ΦC31 att sites flanks the resistance cassette, allowing its unidirectional excision by electroporation of the ΦC31 integrase helper plasmid pJE1817. (C) Plasmid maps of SAGE plasmids used for efficiency experiments in (D). (D) Transformation efficiency when the poly-attP target plasmid pGW60 is transformed with or without an integrase-expressing helper plasmid or when the positive control plasmid is transformed. Control plasmids are as follows: pJE354 (SBW25, TBS10, and Gpo1), pEYF2K (CGA009), or pSET152 (RHA1). Error bars indicate the two-sided SD in three or more biological replicates. Dots indicate individual samples. The accuracy of integration represents the fraction of colonies in which pGW60 recombined into the poly-attB cassette rather than a pseudo-att site, as determined by PCR. With the exception of BT1 in Gpo1 and RV/F370 in RHA1 (which used 12, 20, and 18 samples for screening, respectively), integration accuracy represents the fraction of 24 samples with colony PCR screening results indicating insertion at the intended attB site. n.d. indicates samples not assayed by PCR because of low numbers of transformants across plate replicates. CFU, colony-forming units.