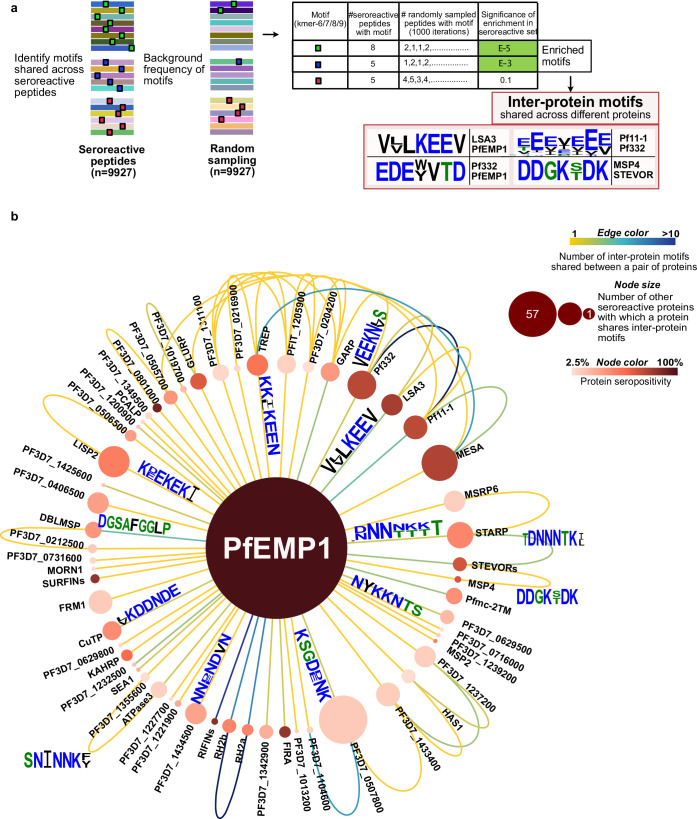
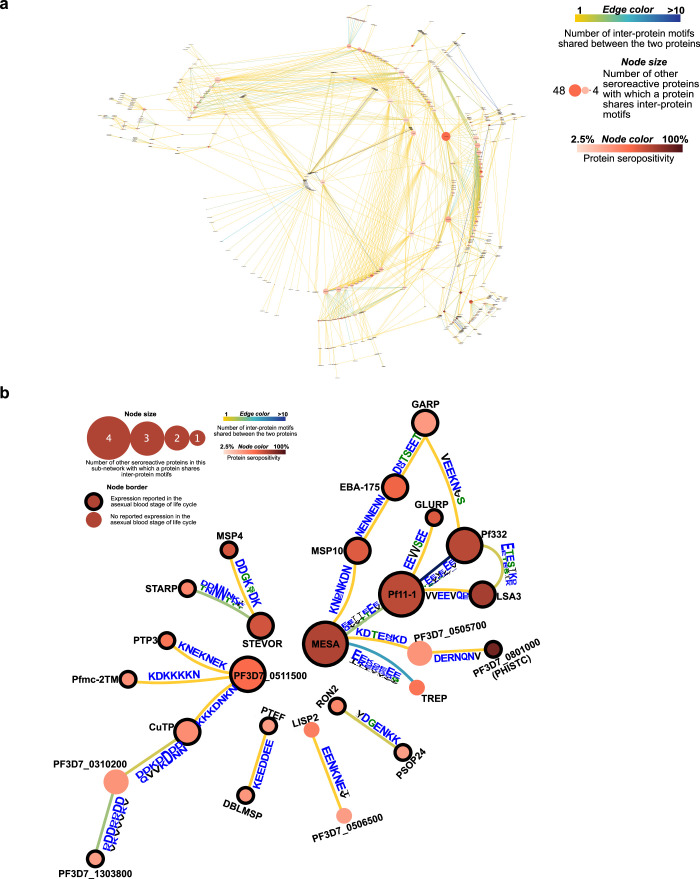
Figure 6. Extensive sharing of motifs observed among seroreactive proteins, with the most shared with PfEMP1 family.
(a) Pipeline to identify inter-protein motifs (6-9aa) significantly enriched (FDR <0.001) in seroreactive peptides from different seroreactive proteins (different colors) over background. Background for each motif was estimated based on the number of random peptides possessing the motif in 1000 random samplings of 9927 peptides. Examples of inter-protein motifs and seroreactive proteins sharing them are also shown. (b) Network of PfEMP1 sharing inter-protein motifs with other seroreactive proteins based on 7-aa motifs with up to two conservative substitutions. PfEMP1 shared inter-protein motifs with the greatest number of other seroreactive proteins.
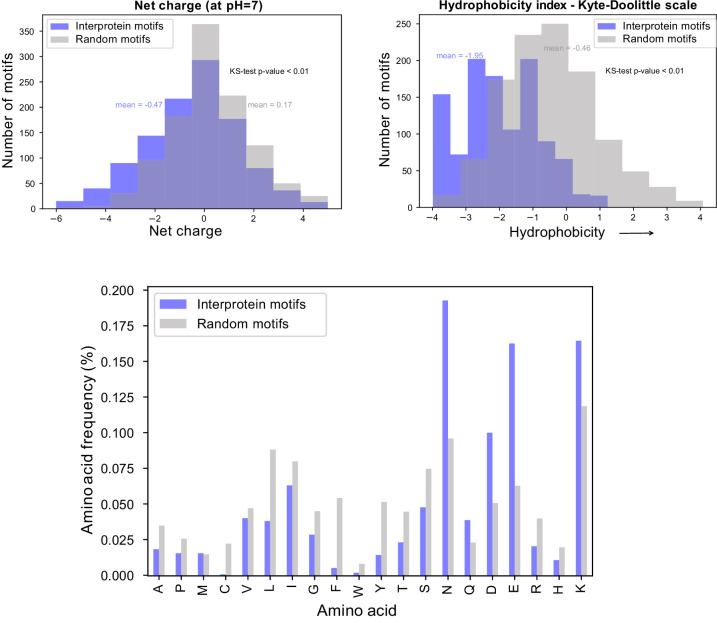
Figure 6—figure supplement 1. Biochemical characteristics of inter-protein motifs.
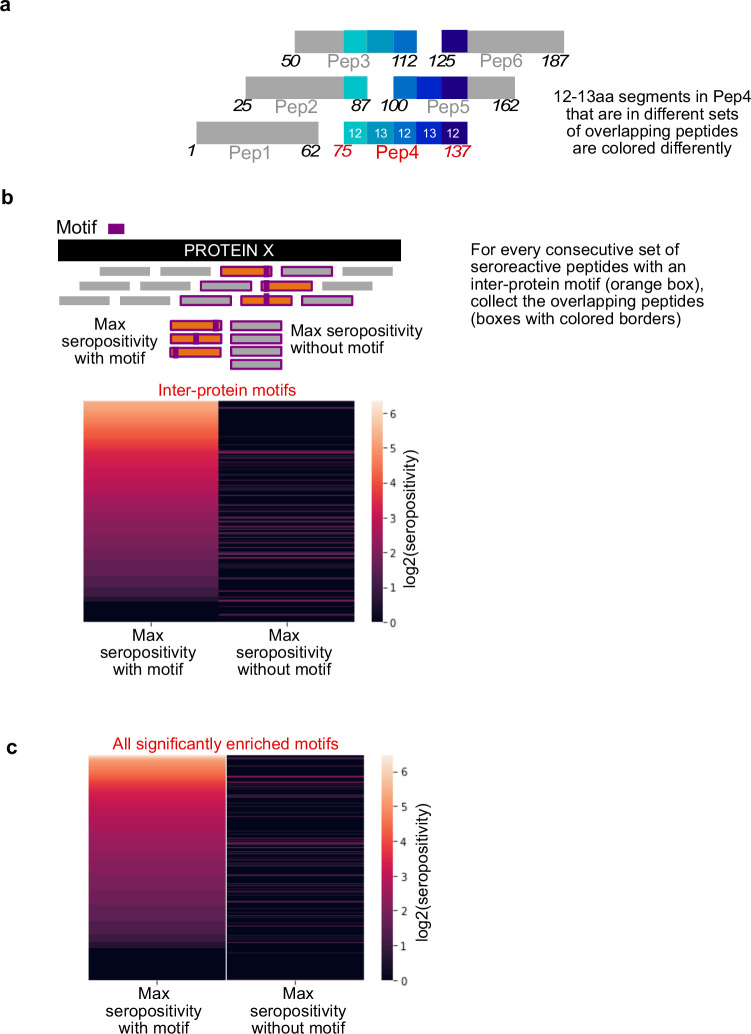
Figure 6—figure supplement 2. Inter-protein motifs are associated with seroreactivity.
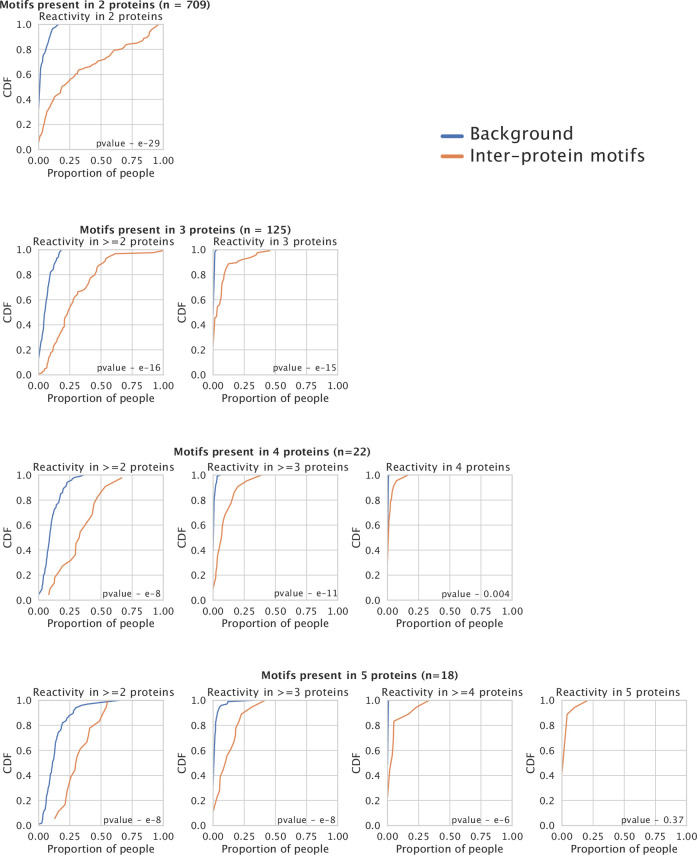
Figure 6—figure supplement 3. Co-occurrence of reactivity to peptides containing inter-protein motifs from different proteins within individuals.
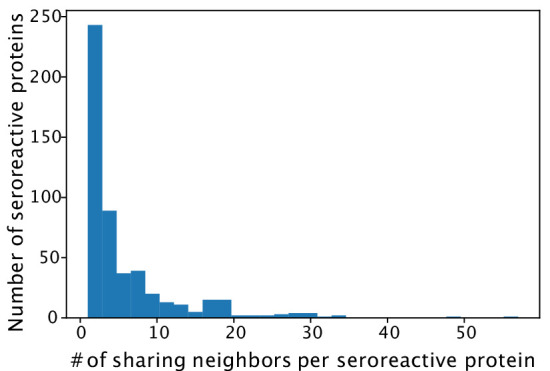
Figure 6—figure supplement 4. Histogram of number of other seroreactive proteins with which a seroreactive protein shares inter-protein motifs.