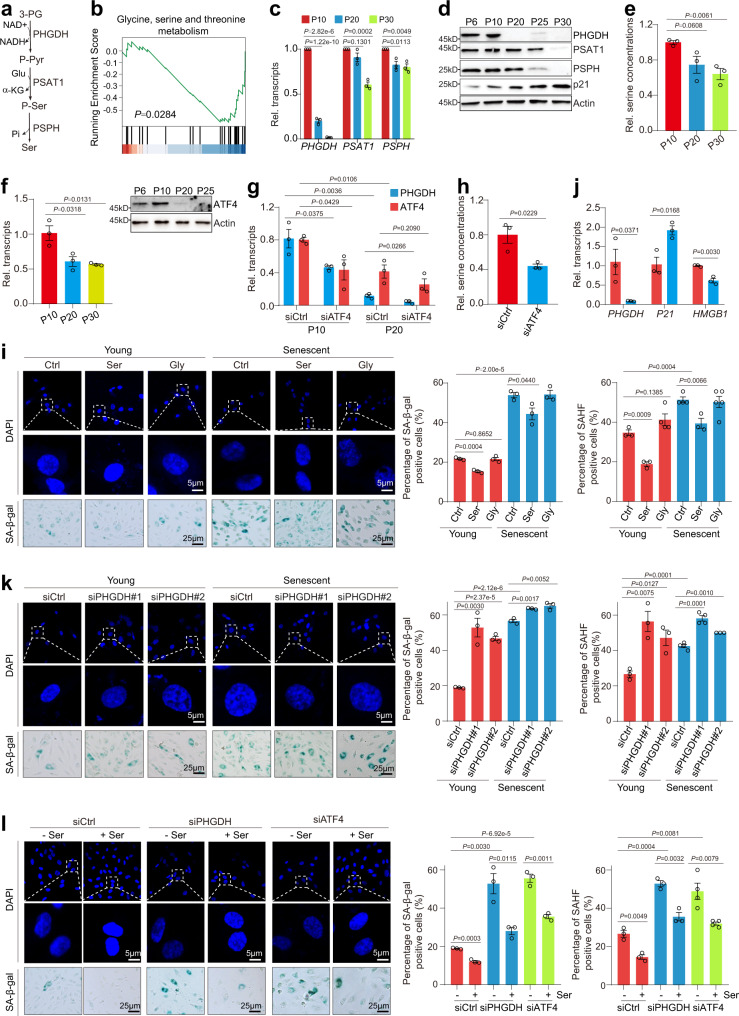
Fig. 1. PHGDH-mediated serine synthesis pathway prevents premature HUVECs senescence.
a Diagram illustrating the de novo serine biosynthesis pathway derived from glycolysis. 3-PG, 3-phosphoglycerate; P-Pyr, 3-phosphohydroxypyruvate; P-Ser, phosphoserine; Ser, serine. b GSEA profiles showing senescence-regulated genes were significantly enriched in glycine, serine and threonine metabolism. Analysis of PHGDH, PSAT1 and PSPH expression in different passages of HUVECs by RT-qPCR (c) and immunoblots (d). e Analysis of the relative intracellular serine levels in different passages of HUVEC cells (P10, P20 and P30) when cultured in serine-free medium. f Analysis of ATF4 expression in HUVECs during senescence by RT-qPCR and immunoblots. g RT-qPCR analysis of ATF4 and PHGDH transcription in siCtrl and siATF4 young (P10) and senescent (P20) HUVECs. h Analysis of the relative intracellular serine levels in siCtrl and siATF4 HUVECs when cultured in serine-free medium. i Effect of serine (Ser) and glycine (Gly) on HUVECs senescence as determined by SAHF formation (DAPI staining) and SA-β-gal staining. HUVECs (young, P10; senescent, P20) were cultured in serine-free medium and then treated with 300 μM serine or glycine for 48 h. Right panel: Quantification of the percentage of SAHF-positive and SA-β-gal positive cells. j RT-qPCR analysis of PHGDH, P21 and HMGB1 transcription in siCtrl and siPHGDH HUVECs when cultured in serine-free medium. k Effect of PHGDH knockdown on the senescence of HUVECs (young, P10; senescent, P20) when cultured in serine-free medium as determined by SAHF formation (DAPI staining) and SA-β-gal staining. Right panel: Quantification of the number of SAHF-positive and SA-β-gal positive HUVECs. l Effect of serine (Ser) on HUVECs senescence in control (siCtrl), PHGDH knockdown (siPHGDH) and ATF4-knockdown (siATF4) HUVECs. Cells were cultured in serine-free medium and then treated with 300 μM serine for 24 h. For h, j, l, HUVECs (P10) were used in the experiments. For c, e–j, data represent means ± SE; n = 3 independent experiments. For i, j, n = 4 independent experiments. Two-sided t-tests were used for statistical analysis. For d, a typical example of three biological replicates is shown.