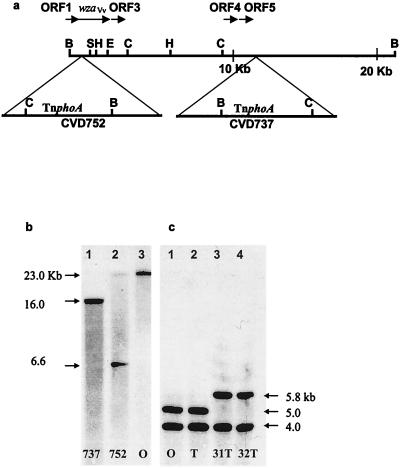
FIG. 1.
Analysis of the V. vulnificus CPS genetic locus. (a) Restriction map for ClaI (C), HindIII (H), SphI (S), and EcoRI (E) sites within the 23-kb BamHI (B) fragment of the V. vulnificus CPS genetic locus. Relative placement and direction of transcription (horizontal arrows) of ORFs and wzaVv are shown. Location of 7.7-kb TnphoA (with a BamHI site at 5 kb) in V. vulnificus CVD737 and CVD752 is indicated by triangles. (b) Southern analysis for BamHI restriction digests of the following V. vulnificus strains: CVD737, lane 1; CVD752, lane 2; and M06-24/O, lane 3. DNA was hybridized with the wzaVv gene probe as described in text. Approximate size of bands (arrows) was determined by extrapolation from known size markers of lambda DNA HindIII digests. CVD737 digests probed with wzaVv show a single band of ca. 16 kb as a product of the new BamHI site introduced 11 kb downstream from the 5′ BamHI site with an additional 5 kb from TnphoA. The insertion in CVD752 occurred within wzaVv 1.6 kb from the 5′ BamHI site to produce 2 bands of ca. 6.6 and 23.4 kb. (c) HindIII restriction digests of chromosomal DNA from the following V. vulnificus strains are shown: lane 1, M06-24/O; lane 2, M06-24/T; lane 3, M06-24/31T; and lane 4, M06-24/32T. DNA was hybridized with a wzaVv probe as described in the text, and the approximate sizes of bands, as determined by comparison to known markers of lambda DNA HindIII digests, are indicated by arrows.