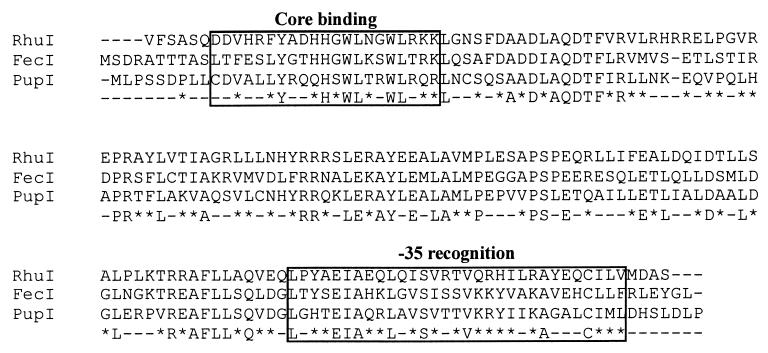
FIG. 3.
Comparison of the amino acid sequence of RhuI to those of two Fur-regulated ECF sigma factors. The predicted amino acid sequence of RhuI was aligned with the amino acid sequences of FecI of E. coli (1) and PupI of P. putida (21) using ClustalW software (http://www.ebi.ac.uk/clustalw/). Regions 2.1 and 4.2, regions important for the function of ECF sigma factors, are boxed, and their functions, core binding and −35 recognition, respectively, are noted. Amino acids are designated by the single-letter code. Dashes in the three sequences represent gaps introduced to maximize alignment. The consensus sequence for the three sequences is shown below the three sequences; in the consensus sequence, positions with amino acid similarity are indicated by asterisks, and positions with nonidentity are designated with dashes. The putative start codon (GTG) of rhuI codes for valine.