Fig. 3 |. Functionally relevant mCRPC enhancers are identified by integrating genetic and epigenetic datasets.
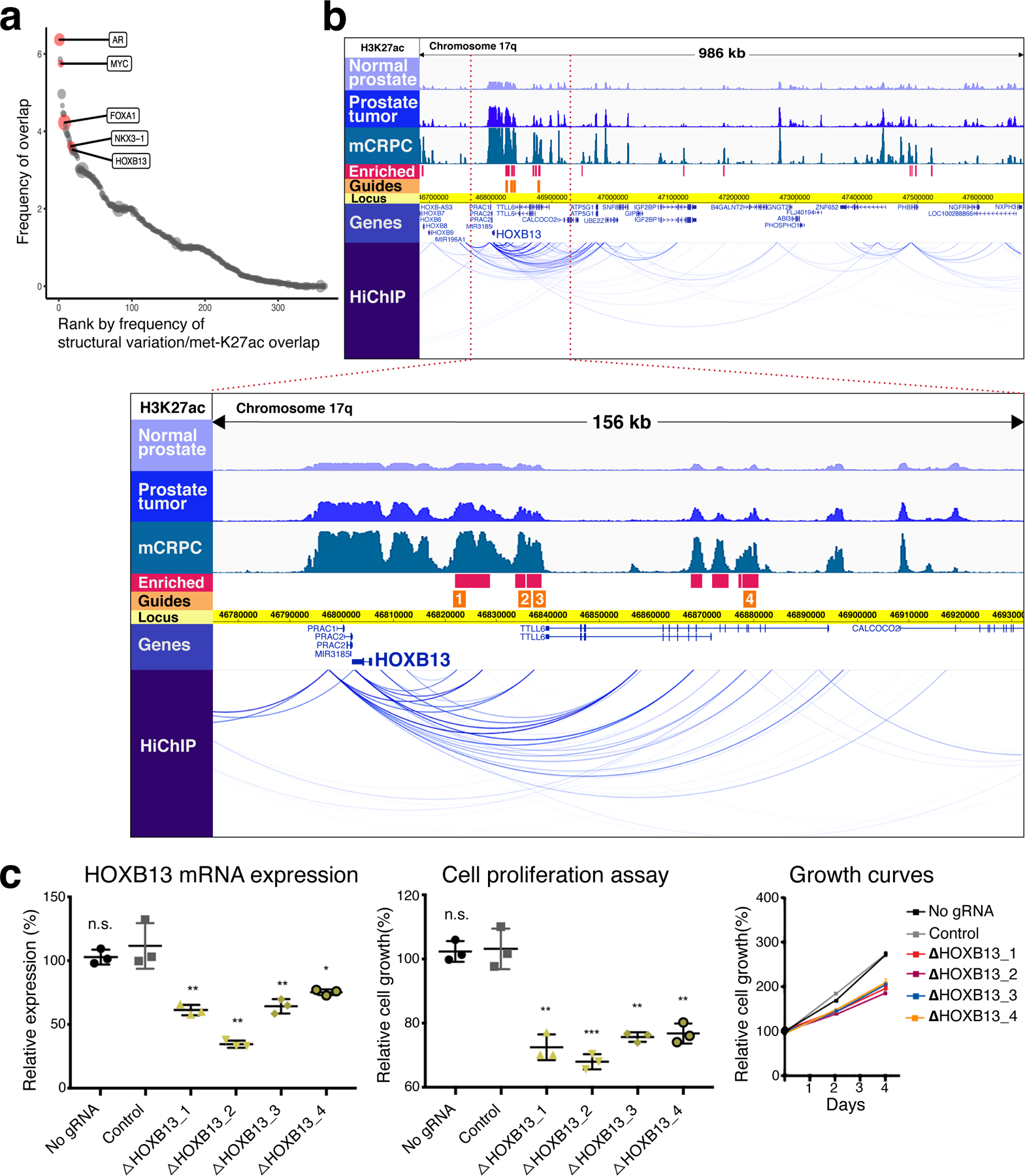
a, Regions of overlap between structural variation in prostate tumors and mCRPC-enriched H3K27ac sites. The size of each circular data point reflects density of mCRPC-enriched H3K27ac signal within the region. Genes of interest falling within specific overlap sites are shown. b, At top, H3K27ac tracks in 986-kb region identified in a containing HOXB13. Intensity of ChIP-seq signal was averaged across all DFCI normal prostate, primary prostate tumor and mCRPC specimens, respectively. HiChIP track depicts chromatin looping in the LNCaP cell line. Blue bars show H3K27ac sites meeting criteria for mCRPC enrichment (met-K27ac). Orange bars depict the locus against which guide RNAs (gRNAs) were designed (Methods). Below, magnification of a 156-kb region (bound by red-dotted lines in the upper picture) where met-K27ac and HiChIP signals were strongest. c, Functional interrogation of candidate metastasis-specific enhancers. Left, LNCaP HOXB13 expression in controls (no gRNA and gRNA targeting unrelated gene HPRT1) and after transduction with each gRNA depicted in b. Middle and right, LNCaP cell proliferation over the course of four days. Each shape represents an independent experiment, center line indicates mean, error bars indicate ± s.d. Using Student’s t-test, two-sided: n.s., not significant; *P < 0.05; **P < 0.01, ***P < 0.001.
