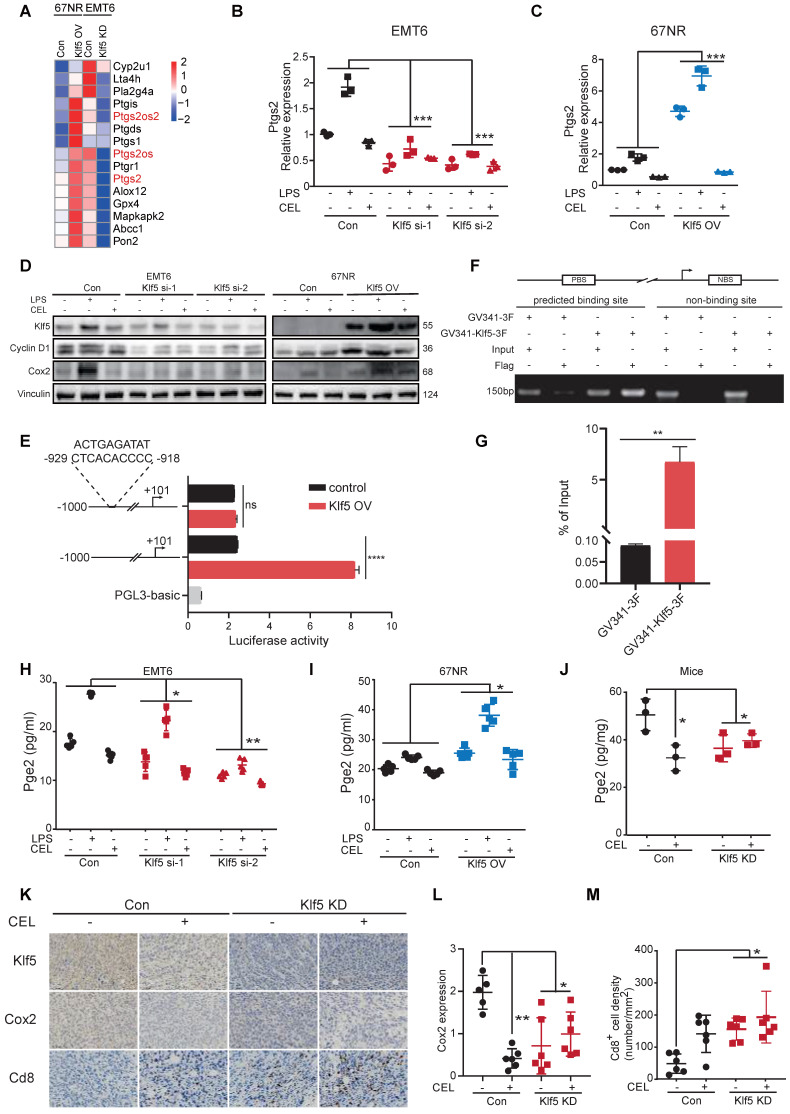
Figure 3.
Klf5 is necessary for tumor-intrinsic PGE2 generation. (A) Heatmap of differentially expressed genes involved in arachidonic acid metabolism from both Klf5-deficient or control EMT6 and Klf5-overexpressing or control 67NR tumors. (B) Relative Ptgs2 (encoded COX2) mRNA expression in control and Klf5-deficient EMT6 cells in the presence or absence of LPS (1 μg/mL) or celecoxib (CEL) (50 μM). (C) Relative Ptgs2 (encoded COX2) mRNA expression in control and Klf5-overexpressing 67NR cells treated with or without LPS (1 μg/mL) or celecoxib (CEL) (50 μM). (D) Western blot analysis of Klf5, CyclinD1 and Cox2 in both Klf5-deficient or control EMT6 and Klf5-overexpressing or control 67NR cells following treatment with or without LPS (1 μg/mL) or CEL (50 μM) for 24 h. (E) HEK293T cells were cotransfected with Ptgs2 (-1000/+101)-luc or Ptgs2mut (-1000/+101)- luc plus GV341-KLF5 or control vector GV341 and the internal control plasmid pRL-TK. (F-G) 67NR wt/Klf5-3F-OV cells were subjected to ChIP assays using anti-Flag magnetic beads. PCR was performed to amplify regions surrounding the putative Klf5 binding region and a nonspecific Klf5 binding region. (H-I) The secreted levels of PGE2 were detected by ELISA in both Klf5-deficient or control EMT6 and Klf5-overexpressing or control 67NR cells following treatment with or without LPS (1 μg/mL) or CEL (50 μM) for 24 h. (J) Pge2 levels were evaluated in Klf5-deficient versus control EMT6 tumors receiving daily CEL (30 mg/kg) treatment for 1 week (n = 3). (K-M) The Klf5, Cox2 and Cd8 levels were quantified by ImageJ after staining with specific antibodies in paraffin-embedded tissues obtained from Klf5-deficient versus control EMT6 tumors. Representative images of Klf5, Cox2 and Cd8 (K). Cox2 expression was quantified in (L). The level of Cd8+ cells was quantified in (M). Scale bar equals 50 μm. The data are represented as means ± SD. n ≥ 5 for mice in each group. (*p < 0.05; **, p < 0.01; ***, p < 0.001 significant vs. control; one-way or two-way ANOVA).