Fig. 5.
Lorlatinib regulates melanoma susceptibility to ferroptosis and PI3K/AKT/mTOR pathway through IGF1R.
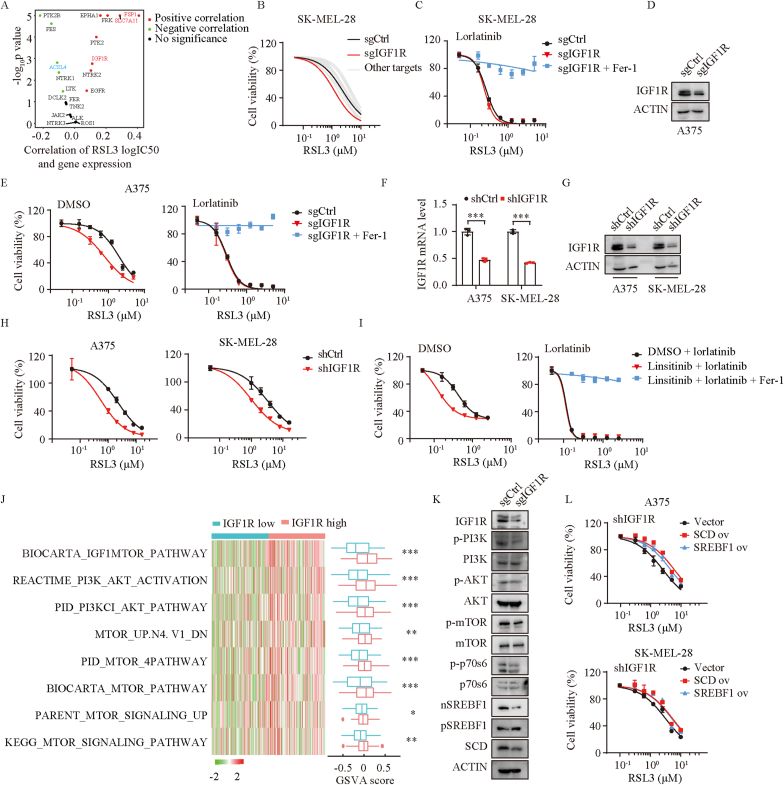
(A) The correlation between logIC50 of RSL3 and gene expression of lorlatinib targets. Red dots, significant positive correlation; blue dots, significant negative correlation; black dots, no significance. (B) Dose response of RSL3-induced death of SK-MEL-28 cells transfected with control sgRNA or sgRNA of lorlatinib targets. SK-MEL-28 cells was sensitive to RSL3 the most when IGF1R was deficient. (C) Dose response of RSL3-induced death of sgCtrl and sgIGF1R SK-MEL-28 cells in the presence of lorlatinib for 6 h. (D) IGF1R protein levels were quantified by western blotting in control (sgCtrl) and IGF1R deficient (sgIGF1R) cells. (E) Dose response of RSL3-induced death of sgCtrl and sgIGF1R A375 cells in the presence of DMSO or lorlatinib (5 μM) for 6 h. (F) IGF1R knock down efficiency in A375 and SK-MEIL-28 cells assessed by real-time PCR (F) and western blotting (G). (H) Dose response of RSL3-induced death of shCtrl and shIGF1R cells. (I) Dose response of RSL3-induced death of DMSO or IGF1R inhibitor (linsitinib) treated- A375 cells in the absence or presence of lorlatinib for 6 h. (J) Gene set variation analysis (GSVA) of TCGA-SKCM segregated by IGF1R expression. High IGF1R group demonstrated significant activation of PI3K/AKT/mTOR related pathways. (K) Western blotting analysis of the indicated proteins in sgCtrl and sgIGF1R A375 cells. (L) Dose response of RSL3-induced death of shIGF1R A375 (upper) and SK-MEL-28 (lower) cells overexpressed vector, SCD or SREBF1. P values were calculated using two-tailed unpaired Student's t-test in F, J. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)