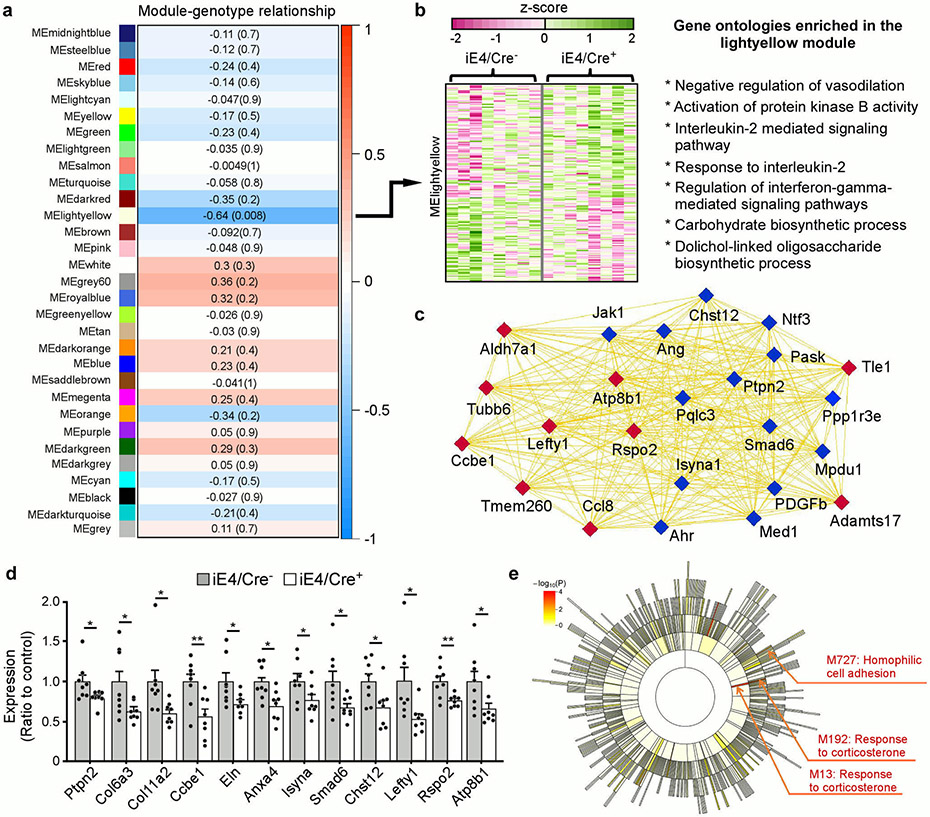
Extended Data Fig. 4. Peripheral apoE4 expression is associated with gene expression profiles consistent with reduced vascular functions and compromised energy homeostasis.
Brain cortical tissues from iE4/Cre mice (Cre−, n=8; Cre+, n=8) at 12-13 month of age were subjected to RNA-Sequencing. a, Module-genotype correlation. Each rectangle represents a module and selected modules are shown. The number in the front of each module is the correlation coefficient (r) between the module eigengene to genotype; the correlation p-value is in the parentheses. Red represents positive correlation to iE4/Cre+ and blue represents negative correlation to iE4/Cre+. b, Heatmaps of genes within the lightyellow module and selected gene ontologies enriched in the module. c, Interaction of genes involved in the gene ontologies (blue nodes) and hub genes (red nodes) in the lightyellow module. The thickness of the lines represents the strength of gene-gene connection. d, Expression of collagen family members (i.e., Col6a3, Col11a2), collagen and calcium binding EGF domain-containing protein 1 (Ccbe1), elastin (Eln), annexin A4 (Anxa4), and genes involved in carbohydrate biosynthetic process as well as the hub genes (Lefty1, Rspo2 and Atp8b1) in the lightyellow module shown in (c) were validated in iE4/Cre mice (Cre−, n=8; Cre+, n=8) by real-time PCR analysis. Data expressed as mean ± s.e.m. Ptpn2 (*, P=0.042); Col6a3 (*, P=0.036); Col1a2 (*, P=0.027); Ccbe1 (**, P=0.006); Eln (*, P=0.04); Anxa4 (*, P=0.014); lsyna (*, P=0.047); Smad6 (*, P=0.041); Chst12 (*, P=0.028); Lefty1 (*, P=0.021); Rspo2 (**, P=0.006); Atp8b1 (*, P=0.031), two-tailed Student's t-test. e, Sunburst plot showing the module hierarchical structure relationship. Each rectangular block denotes a module. A total of 1002 modules were identified in the E4 network, with module size ranging from 10 to 960. The color intensity denotes the FDR adjusted P value significance of GSEA based enrichment for differential expression signals. Functional annotations are highlighted for modules overrepresented with GO/pathway genes (red text for positive GSEA enrichment score with regard to differential expression signal).