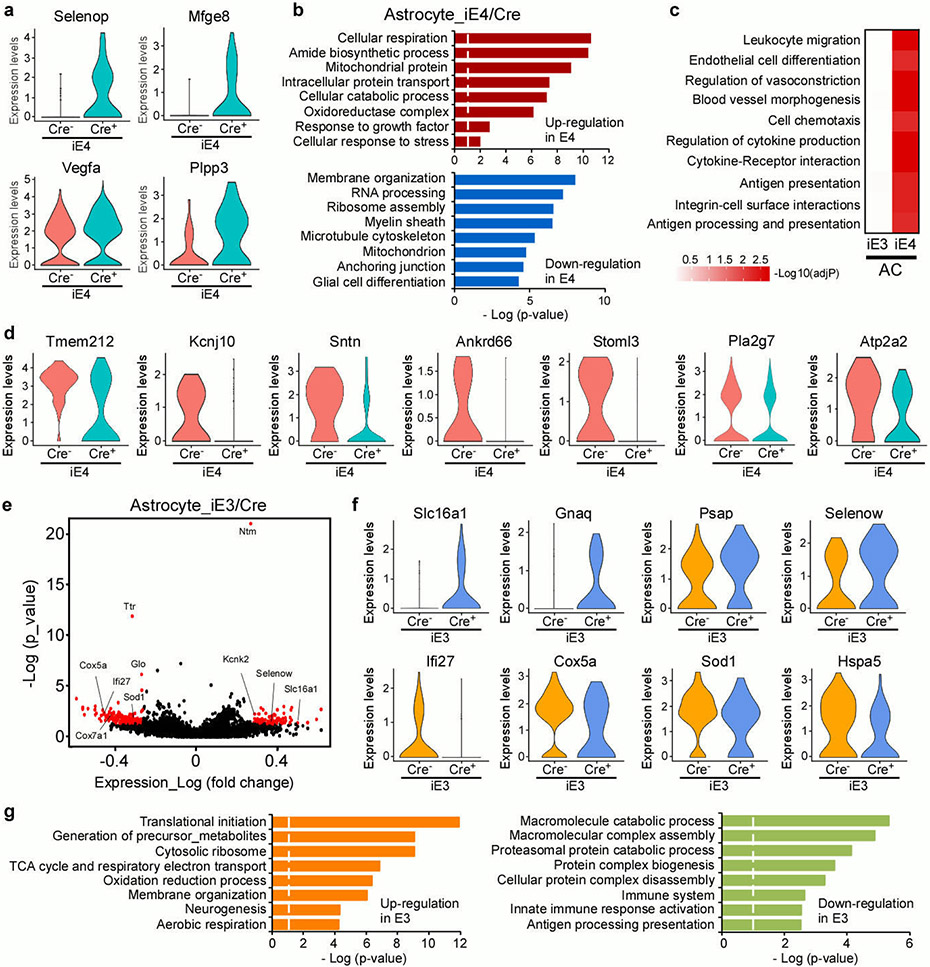
Extended Data Fig. 6. Brain transcriptional changes in astrocytes influenced by peripheral apoE isoform expression.
a, Violin plots showing the mean and variance differences between iE4/Cre− and iE4/Cre+ astrocytes for genes regulating oxidative stress, lipid metabolism, and hypoxia/stress responses (Selenop, Mfge8, Plpp3, and Vegfa). b, Gene ontology (GO) enrichment analysis for genes upregulated (red) or down-regulated (blue) in astrocytes from iE4/Cre+ mice compared to iE4/Cre− mice. c, Top gene ontology and canonical pathways enriched for DEGs in the astrocytes from iE3/Cre and iE4/Cre mice were identified by gene set enrichment analysis (GSEA). d, Violin plots showing the mean and variance differences between iE4/Cre− and iE4/Cre+ astrocytes for genes regulating astrocyte endfeet (Tmem212, Kcnj10, Sntn, Ankrd66, Stoml3, Pla2g7, Atp2a2). e, Volcano plot depicting up- and down-regulated genes in astrocytic cell populations (AC1, AC2, AC3) in iE3/Cre+ mice compared to iE3/Cre− control mice. Genes significant at the P value ≤ 0.05 and fold change ≥ 1.2 are denoted in red. f, Violin plots showing the mean and variance differences between iE3/Cre− and iE3/Cre+ astrocytes for genes regulating lactate homeostasis (Slc16a1), cellular protection (Psap and Selenow), immune response (Ifi27, Cox5a), and cellular senescence/stress response (Sod1 and Hspa5). g, Gene ontology enrichment analysis for genes up-regulated (orange) or down-regulated (green) in astrocytes from iE3/Cre+ mice compared to iE3/Cre− control mice. Limma’s moderate t-test statistics were used to rank the transcriptome-wide gene-phenotype association.