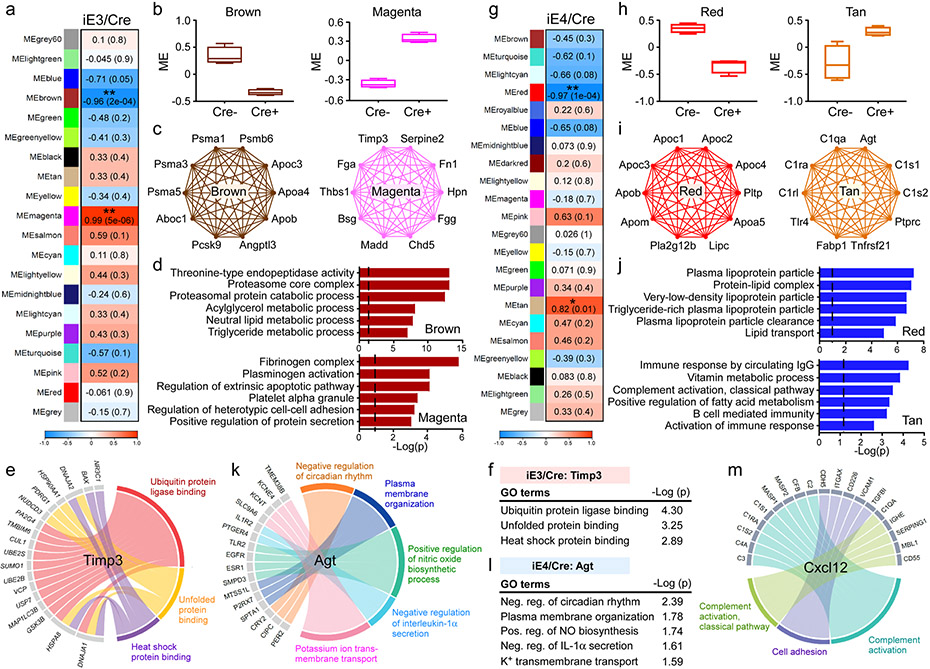
Fig. 5 ∣. ApoE isoform-specific changes in plasma protein network and the association with vascular related pathway.
Plasma from iE3/Cre (a-f) mice (Cre−, n=4; Cre+, n=4), and iE4/Cre (g-m) mice (Cre−, n=4; Cre+, n=4) were subjected to proteomics analysis. The protein modules (MEs) associated with APOE3 or APOE4 genotype were identified by WGCNA analysis. a, g, Module-genotype correlation. Numbers in the table indicate the correlations of the corresponding module eigengenes and genotype, with the P values shown in parentheses. The red color represents positive correlation and blue represents negative correlation to APOE genotype. b, h, Modules that are significantly correlated with APOE3 (b) or APOE4 (h) genotype. The box in the plots displays 25th and 75th percentile values. The center line represents the median and the whiskers show minimum and maximum values. c, i, Network plots of the proteins with high intramodular connectivity in the brown and magenta modules in the iE3/Cre group (c) and red and tan modules in the iE4/Cre group (i). d, j, Pathway analysis using gene ontology (GO) showing the pathways enriched in the protein-sets of the brown and magenta modules in the iE3/Cre group (d), as well as red and tan modules in the iE4/Cre group (j). Limma’s moderate t-test statistics were used. e, Circos plot showing the associated genes and pathways in the endothelial cell clusters negatively correlated with the changes of plasma Timp3 in the iE3/Cre group. f, Timp3-regulated pathways in the endothelial cell clusters of the iE3/Cre group. k, Circos plot showing the associated genes and pathways in the endothelial cell clusters negatively correlated with the changes of plasma Agt in the iE4/Cre group. l, Agt-regulated pathways in the endothelial cell clusters of the iE4/Cre group. Limma’s moderate t-test statistics were used. m, Circos plot showing the associated proteins and pathways in the plasma correlated with the changes of Cxcl12 in endothelial cell clusters in the iE4/Cre group.