Summary
Opposing effects of interferon-induced transmembrane proteins (IFITMs 1, 2 and 3) on SARS-CoV-2 infection have been reported. The reasons for this are unclear and the role of IFITMs in infection of other human coronaviruses (hCoVs) remains poorly understood. Here, we demonstrate that endogenous expression of IFITM2 and/or IFITM3 is critical for efficient replication of SARS-CoV-1, SARS-CoV-2 and hCoV-OC43 but has little effect on MERS-, NL63-and 229E-hCoVs. In contrast, overexpression of IFITMs inhibits all these hCoVs, and the corresponding spike-containing pseudo-particles, except OC43, which is enhanced by IFITM3. We further demonstrate that overexpression of IFITMs impairs cell surface expression of ACE2 representing the entry receptor of SARS-CoVs and hCoV-NL63 but not hCoV-OC43. Our results explain the inhibitory effects of artificial IFITM overexpression on ACE2-tropic SARS-CoVs and show that three hCoVs, including major causative agents of severe respiratory disease, hijack IFITMs for efficient infection of human cells.
Subject areas: Protein, Immunity, Virology
Graphical abstract
Highlights
-
•
Three of five human β-coronaviruses hijack IFITMs for efficient infection
-
•
IFITMs are critical for efficient replication of the two causative agents of SARS
-
•
Overexpression of IFITMs inhibits SARS-CoVs by suppressing ACE2 cell surface expression
-
•
Inhibitory membrane modulation but not S interaction is conserved between men and mice
Protein; Immunity; Virology
Introduction
As a consequence of millions of years of an evolutionary arms race with pathogens, humans have evolved elaborate antiviral defense mechanisms. Many human genes encode structurally and functionally diverse antiviral effectors, referred to as ‘restriction factors’ (RFs). Some of them are constitutively expressed in cells to prevent infection but most are induced on innate immune activation, e.g., by interferons (IFNs). RFs typically target conserved viral features or cellular dependency factors and are thus active against a broad spectrum of viral pathogens.1,2,3 However, viruses demonstrate a striking ability to adapt to their hosts and evade or counteract antiviral mechanisms.4,5 Accumulating evidence shows that some of the most successful viral pathogens may even exploit otherwise antiviral cellular factors for efficient infection and replication.6
One group of broad-spectrum antiviral factors is a member of the IFN-induced transmembrane (IFITM) family.7,8,9 IFITMs are typically small (125–133 amino acids) transmembrane proteins, which have been reported to inhibit entry of retro-, orthomyxo-, paramyxo-, flavi-, filo-, rhabdo-, influenza A and coronaviruses (CoVs).7,8,10,11 Humans encode five IFITM proteins and at least three of them (1, 2, and 3) can display antiviral activity. It is believed that IFITMs do not restrict viral entry by targeting specific viral compounds but by modulating membrane composition, fluidity and curvature.12,13,14 Viral and cellular membranes differ in their lipid composition.15,16 Viral membranes require strong curvature because viruses are much smaller than their target cells and strong bending is required for virion-cell fusion. Thus, it has been suggested that IFITMs increase membrane “stiffness” to prevent efficient virion-cell fusion.9,12,13,14,17
Although IFITMs are best known for their broad antiviral activity, accumulating evidence suggests that some viruses are able to exploit them for infection. An elegant study by Zhao et al. showed that the common cold coronavirus (ccCoV) OC43 utilizes IFITM3 for efficient entry into human hepatoma cell lines.18 In contrast, inhibitory effects of IFITM overexpression have been reported for the three highly pathogenic human coronaviruses SARS-CoV-1, SARS-CoV-2 and MERS-CoV.19,20,21 We previously confirmed that overexpression of IFITMs inhibits infection by spike(S)-containing lentiviral and vesicular-stomatitis virus(VSV)-based pseudo-particles, as well as genuine SARS-CoV-2.22 In contrast, however, endogenous expression of IFITMs was essential for efficient replication of early pandemic SARS-CoV-2,22 and all five variants of concern known to date.23 Unlike the broad inhibitory effects of IFITMs on virion-cell fusion, enhancement of SARS-CoV-2 infection most likely involves specific interactions between the viral spike (S) protein and IFITMs.22
The reasons for the reported opposing effects of overexpressed and endogenous IFITMs remained elusive. In addition, many results on effects of IFITM proteins on hCoVs infection were obtained using S-containing pseudo-virions and overexpression assays. Thus, whether endogenously expressed IFITMs act as enhancers or inhibitors of most genuine hCoVs, including their most virulent representatives SARS-CoV-1 and MERS-CoV remained unclear. Here, we analyzed the effects of overexpressed and endogenous IFITMs on S-mediated viral pseudo-particle and genuine virus infection for six of the seven hCoVs. We demonstrate that endogenously expressed IFITMs are critical for effective replication of SARS-CoV-1, SARS-CoV-2 and hCoV-OC43. In contrast, overexpression of IFITMs essentially inhibited infection of all hCoVs and S-containing viral pseudo-particles by both altering membrane features and impairing cell surface expression of ACE2, the receptor of SARS-CoV-1, SARS-CoV-2 and hCoV-NL63. The only exception was OC43 S-mediated infection, which does not require ACE2 and was generally enhanced by IFITM3. Our results explain why studies analyzing overexpressed or endogenous IFITMs yielded opposing results and demonstrate that IFITMs are critical cofactors for efficient infection of both SARS coronaviruses.
Results
Human hepatocytes are highly susceptible to infection by common cold coronaviruses
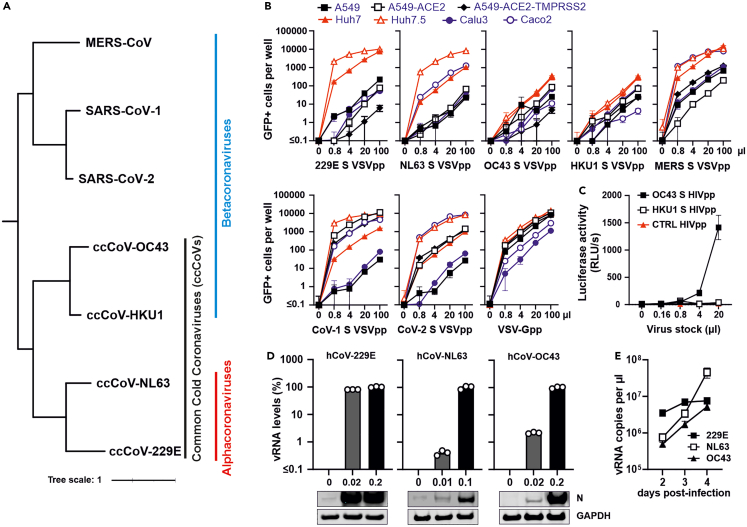
To establish cellular models for S-mediated pseudo-particle (pp) infection and genuine hCoVs, we first generated single-cycle VSV particles encoding GFP instead of the open reading frame for the viral glycoprotein (VSV(GFP)ΔG) pseudo-typed with the S proteins of all seven human CoVs (Figure 1A) and infected a panel of seven cell lines commonly used to study coronavirus infection. The human hepatoma-derived Huh-7 cell line was efficiently infected by Vesicular stomatitis virus pseudoparticles (VSVpp) containing the hCoV-229E, hCoV-NL63, MERS, CoV-1 and CoV-2 S proteins or the VSV-G control (Figure 1B). Huh-7 cells are commonly used as model for primary human hepatocytes and show significant similarities in gene expression profiles.24,25 The Huh7.5 derivate, which has a defect in the retinoic-acid-inducible gene-I (RIG-I) pathway,26 was even more susceptible to S-mediated VSVpp infection. Infection of other cell lines was more variable and often less efficient. The epithelial Caco-2 cell line derived from a colon carcinoma, promoted efficient infection by hCoV-NL63, MERS and CoV-2 S, whereas A549-ACE2 and A549-ACE2-TMPRSS2 cells mediated infection by CoV-1 and CoV-2 S proteins (Figure 1B). In comparison, parental A549 cells and Calu-3 cells showed low susceptibility to S-mediated VSVpp infection. Altogether, VSVpp provided a suitable system for S-mediated infection for five of the seven hCoVs (Figure 1B). However, VSV-based pseudo-particles carrying the hCoV-OC43 and-HKU1 S proteins were generally poorly infectious, possibly because of inefficient virion incorporation of these spike proteins. To further expand the panel, we generated lentiviral pseudo-particles (HIVpp) carrying the hCoV-OC43 and-HKU1 S proteins by transfection of HEK293T cells with vectors expressing the S proteins together with a pNL1_HIV-1_NL4-3-Δenv-fluc construct.22 This allowed significant infection mediated by the hCoV-OC43 S (Figure 1C). In contrast, the titers of HIVpp containing hCoV-HKU1 S were too low for meaningful analyses. Notably, it has been previously reported that propagation of hCoV-HKU1 in immortalized cells is highly challenging.27
Figure 1.
Susceptibility of various cell types to infection by spike-containing viral pseudo-particles and genuine human coronaviruses
(A) Phylogenetic tree of human coronaviruses. Distance-based relationship inference based on representative full-genome nucleotide sequences of the indicated human coronaviruses.
(B) Automatic quantification of infection events of A549, A549-ACE-2, A549-ACE-2-TMPRSS2, Huh7, Huh7.5, Calu3 and Caco2 cells transduced with different quantities of VSVΔG-GFP stocks pseudo-typed with hCoV-229E,-NL63,-OC43, HKU1, MERS, CoV-1 and CoV-2 spike proteins or VSV-G for control. Cytation was performed at 24 h post-infection.
(C) Lentiviral particles containing hCoV-OC43 or-HKU1 S proteins were generated by cotransfection of HEK293T cells with spike expression plasmids and a pNL1_HIV-1_NL4-3-Δenv-fluc construct. Huh7 cells were infected with the indicated amounts of HIVpp and luciferase activity was measured at 48 h post-infection.
(D) Huh7 cells were infected with genuine hCoV-229E, hCoV-NL63 and hCoV-OC43 at the indicated MOIs. Viral RNA levels in cellular extracts were determined by qRT-PCR 48 h post-infection. The lower panel shows immunoblots of the corresponding whole cell lysates stained with anti-229E N,-NL63 N or-OC43 N and anti-GAPDH.
(E) Huh7 cells were infected with hCoV-229E (MOI 0.01), hCoV-NL63 (MOI 0.05) and hCoV-OC43 (MOI 0.01). Viral RNA in supernatants harvested at 2, 3 and 4 days post-infection was determined by qRT-PCR. Bars represent the mean of three independent experiments (±SEM).
For genuine viruses, we initially focused on the seasonal coronaviruses because we had already established Calu-3 cells as suitable system for infection by highly pathogenic CoVs.22,28 In agreement with the results obtained using S-containing pseudo-particles, Huh7 cells supported replication of three of the four seasonal viruses: hCoV-OC43 (Organ Culture 43), which belongs to the β-coronaviruses (just like the highly pathogenic SARS- and MERS-CoVs; Figure 1A), and the two α-coronaviruses hCoV-NL63 (Netherland 63) and −229E (Figures 1D and 1E). Altogether, our results showed that Huh7 cells are suitable to study S-mediated infection by six of the seven human coronaviruses.
IFITM overexpression inhibits hCoV-229E and-NL63 but promotes hCoV-OC43 infection
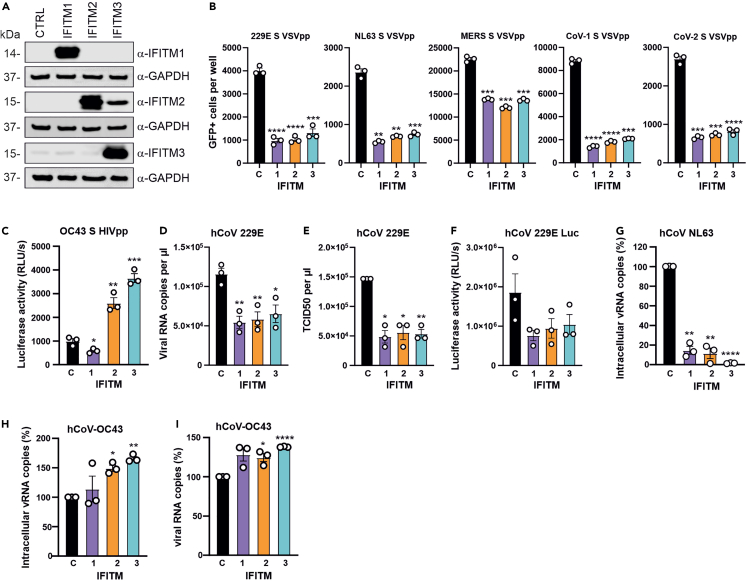
To determine the impact of IFITM overexpression on S-mediated pseudo-particle and genuine ccCoV infection, we transfected Huh7 cells with expression constructs for the three IFITM proteins. Western blot analysis verified that transfection with the respective expression vectors resulted in high levels of IFITM1, 2 and 3 expression (Figure 2A). Overexpression of all three IFITM proteins significantly reduced infection by 229E, NL63, MERS, CoV-1 and CoV-2 S VSVpp (Figure 2B). In contrast, IFITM2 and IFITM3 increased OC43 S-mediated HIVpp infection by ∼3-fold (Figure 2C). Notably, we have previously shown that HIVpp containing the SARS-CoV-2 S are inhibited by IFITM overexpression.22 Thus, the enhancing effect was specific for OC43 S-mediated infection and not dependent on the carrier particle.
Figure 2.
Effect of IFITM overexpression on S-mediated VSVpp or HIVpp and ccCoV infection
(A) Huh7 cells were transiently transfected with plasmids expressing indicated the IFITM proteins or an empty control (CTRL) vector. Shown are immunoblots of whole cell lysates stained with anti-IFITM1, anti-IFITM2, anti-IFITM3 and anti-GAPDH. Note that the α-IFITM2 Ab shows some cross-reactivity with IFITM3.
(Band C) Huh7 cells transfected with vectors expressing the indicated IFITM were infected with (B) VSVpp containing the hCoV-229E,-NL63, MERS CoV, SARS CoV-1 or SARS CoV-2 S proteins or (C) HIVpp carrying the hCoV-OC43 S. GFP+ cells and luciferase activities were determined 24h and 48h post-infection, respectively.
(D and E). Huh7 cells transfected with vectors expressing indicated IFITM proteins were infected with hCoV-229E at an MOI 0.01. Culture supernatants were harvested at 24h post-infection and (D) viral RNA or (E) infectious virus yields were measured by qRT-PCR and TCID50 assay, respectively.
(F) Huh7 cells overexpressing the indicated IFITMs were infected with Renilla hCoV-229E (MOI 0.01) and luciferase activity was measured 24h later.
(G) Huh7 cells overexpressing the indicated IFITM proteins were infected with hCoV-NL63 (MOI 0.05). Cell lysates were harvested at 48h post-infection and intracellular hCoV-NL63 RNA levels were determined by qRT-PCR.
(H and I) Huh7 cells transiently expressing the indicated IFITM proteins were infected with hCoV-OC43 (MOI 0.01). Culture supernatants and whole cell lysates were harvested for qRT-PCR at 48h post-infection to determine the viral RNA level in the supernatant (H) or intracellular viral RNA levels (I). Bars in all panels represent the mean of three or four independent experiments (±SEM); ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p > 0.0001.See also Figure S1.
In agreement with the results obtained using 229E S VSVpp, overexpression of all three IFITMs inhibited RNA production (Figure 2D) and infectious virus yields (Figure 2E) in Huh7 cells infected with genuine hCoV-229E. The inhibitory effects of IFITM overexpression was confirmed using a replication-competent version of hCoV-229E expressing a luciferase reporter gene (Figure 2F).29 Overexpression of IFITMs had even stronger inhibitory effects on hCoV-NL63: IFITM1 and IFITM2 reduced viral RNA levels by ∼90% and IFITM3 blocked infection almost entirely (Figure 2G). In contrast, IFITM overexpression moderately increased intracellular RNA levels (Figure 2H) and viral RNA production (Figure 2I) of hCoV-OC43. The relatively modest effect of IFITM3 overexpression is likely because of basal IFITM3 expression levels in Huh7 cells that may already promote hCoV-OC43 infection. Altogether, these results confirmed and expanded previous data showing that IFITM overexpression inhibits S-mediated entry of most hCoVs but has the opposite effect on hCoV-OC43 infection.
Effect of endogenous IFITMs on ccCoV infection
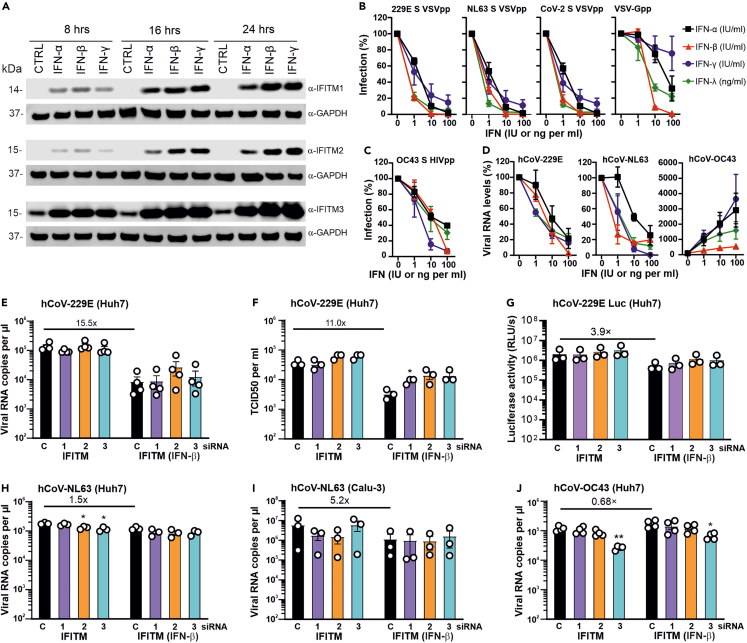
To determine whether IFNs induce expression of IFITMs and an antiviral state in Huh7 cells, we pre-treated these cells with IFN-α, IFN-β and IFN-γ and analyzed them by western blot. Similar to our previous results obtained using Calu-3 cells,22 type I and II IFNs induced efficient expression of all three IFITMs in Huh7 cells (Figure 3A). As expected, IFN treatment efficiently inhibited hCoV-229E, hCoV-NL63 and CoV-2 S-mediated VSVpp infection (Figure 3B), as well as OC43 S-mediated HIVpp infection (Figure 3C). In comparison, IFNs also inhibited genuine hCoV-229E and-NL63 but enhanced replication of hCoV-OC43 (Figure 3D). IFN-α and IFN-γ were most effective and resulted in up to 35-fold higher levels of vRNA production in hCoV-OC43 infected Huh7 cells. Altogether, all S-containing pseudo-particles showed similar susceptibility to IFN inhibition and did not recapitulate the enhancing effect of IFNs on genuine hCoV-OC43 replication. These results suggest that IFN sensitivity of VSVpp and HIVpp is determined by the viral backbone rather than the pseudo-type S-proteins they carry.
Figure 3.
Effect of IFNs and endogenous IFITM expression on ccCoVs
(A) Huh7 cells were stimulated with IFN-α2, IFN-β or IFN-γ for 8h, 16h and 24h. Immunoblot was performed using whole cell lysates stained with anti-IFITM1, anti-IFITM2, anti-IFITM3 and anti-GAPDH.
(B) Huh7 were left untreated or treated with IFN-α, IFN-γ and IFN-λ for 18h and subsequently infected with 229Epp, NL63pp, SARS CoV-2pp or VSV-Gpp. Cytation for GFP+ cells was performed at 24h post-infection.
(C) Huh7 cells were treated with IFNs as described in panel B and infected with HIVpp carrying the hCoV-OC43 S protein. Luciferase activity was determined at 48h post-infection.
(D) Huh7 cells were treated with IFNs and infected with genuine hCoV-229E (MOI 0.01), hCoV-NL63 (MOI 0.05) or hCoV-OC43 (MOI 0.01). Whole cell lysates were collected at 24h post-infection for hCoV-229E and at 48h post-infection for hCoV-NL63 and hCoV-OC43. Intracellular viral RNA levels were determined by qRT-PCR.
(E and F) Huh7 cells were transfected with the indicated siRNAs, left untreated or treated with IFN-β 100U/ml) for 18h starting on the following day and subsequently infected with hCoV-229E (MOI 0.01) after 18h of IFN treatment. Supernatant was harvested 24h post-infection and hCoV-229E RNA and infectious titer quantified by qRT-PCR and TCID50 assay, respectively.
(G) Huh7 cells were treated as described for panel E, infected with Renilla hCoV-229E (MOI 0.01) and luciferase activity was measured 24h later.
(H) Huh7 cells transfected with IFITM targeting or non-targeting siRNA were treated with IFN-β (100 U/ml) or left untreated, infected with hCoV-NL63 (MOI 0.05) and viral RNA levels in the culture supernatants were determined by qRT-PCR two days later.
(I) Calu3 cells transfected with IFITM targeting or non-targeting siRNA were treated with IFN-β (500 U/ml) or left untreated, infected with hCoV-NL63 (MOI 0.05) and viral RNA levels in the culture supernatants were determined by qRT-PCR two days later.
(J) Huh7 cells were treated as described in panel H, infected with hCoV-OC43 and viral RNA levels in the culture supernatants were determined two days later. Bars in all panels represent the mean of three or four independent experiments (±SEM). ∗, p < 0.05; ∗∗, p < 0.01).
To examine the effect of endogenous IFITM expression on S-mediated infection, we performed siRNA knock-down (KD) studies in Huh7 cells. siRNA treatment reduced the levels of IFITM mRNA expression by >90% (Figure S1A) but had only modest enhancing effects on infection by hCoV-229E, hCoV-NL63, MERS, CoV-1 and CoV-2 S VSVpp (Figure S1B). In comparison, silencing of IFITM3 markedly reduced OC43 S-mediated infection of Huh7 and Huh7.5 cells (Figures S1C and S1D). To examine the impact of IFITM depletion on genuine hCoVs, we performed siRNA KD analyses in untreated and IFN-β-treated Huh7 cells. Treatment with siRNAs generally decreased the levels of targeted IFITM mRNAs by about one order of magnitude (Figure S1E). Silencing of endogenous IFITM expression had little effect on viral RNA production (Figure 3E) and infectious virus yields (Figure 3F) in hCoV-229E infected Huh-7 cells. In accordance with this, IFITM KD had little impact on infection by the hCoV-229E luciferase reporter virus (Figure 3G). Similarly, individual silencing of the three IFITM proteins did not significantly affect hCoV-NL63 replication in Huh7 (Figure 3H) and Calu-3 cells (Figure 3I). In contrast, silencing of IFITM3 reduced the levels of viral RNA production in hCoV-OC43 infected Huh7 cells by 70–80% in both untreated and IFN-β-treated Huh7 cells (Figure 3J). Altogether, these results indicate that IFITM overexpression clearly inhibits hCoV-229E and hCoV-NL63, while endogenous expression had little effect. In contrast, IFITM3 considerably enhanced hCoV-OC43 replication under all experimental conditions used.
IFITM2 and IFITM3 are critical for efficient replication of SARS-CoV-1
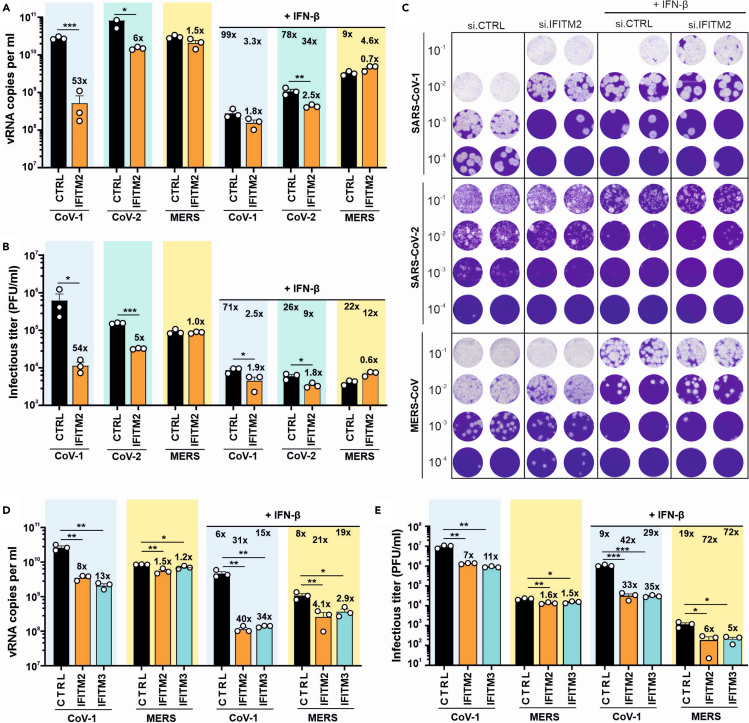
We have shown that efficient SARS-CoV-2 replication in Calu-3 cells is highly dependent on IFITM2 and (to a lesser extent) on IFITM3, while silencing of IFITM1 had only modest effects.22,23 Thus, we asked whether endogenous IFITM2 expression also affects infection of human lung cells by the two most virulent human coronaviruses, SARS-CoV-1 and MERS-CoV. SARS-CoV-1 is closely related to SARS-CoV-2 and infected over 8,000 people with a case fatality rate of about 10% between 2002 and 2004.30 MERS-CoV is only distantly related to SARS-CoV-2 and was first reported in 2012. Since then it has infected over 2,500 humans resulting in 900 deaths. It has previously been established that Calu-3 cells express all three IFITM proteins and are susceptible to infection by highly pathogenic human CoVs.22,28 Our siRNA silencing approach reduces IFITM2 expression by about 90%,22 and significantly impaired SARS-CoV-2 RNA production and infectious virus yields (Figures 4A–4C). Silencing of IFITM2 expression had even stronger effect on SARS-CoV-1 replication and reduced viral RNA production and infectious virus titers by > 50-fold (Figures 4A–4C). In contrast, IFITM2 KD had no significant effect on infection and replication of MERS-CoV. IFN-β treatment resulted in almost complete inhibition of SARS-CoV-1 and SARS-CoV-2, explaining why IFTIM2 siRNA KD had only modest enhancing effects in the presence of IFN-β.
Figure 4.
Endogenous IFITM expression is critical for efficient SARS-CoV replication
(A) Quantification of viral E gene RNA by qRT-PCR in the supernatant of Calu-3 48 h post-infection with SARS-CoV-1, SARS-CoV-2 or 24 h post-infection with MERS-CoV. Cells were transiently transfected with siRNA either control (CTRL) or targeting IFITM2 as indicated. In addition, cells were either treated with IFN-β (250 U/ml for SARS-CoV and 50 U/ml for MERS-CoV) or left untreated. Bars in all panels represent the mean of three independent experiments (±SEM). ∗, p < 0.05; ∗∗; p < 0.01; ∗∗∗, p < 0.001. The numbers directly above the bars indicate fold change compared to cells treated with control (CTRL) siRNA and number below the horizontal lines fold reduction on IFN-β treatment.
(B and C) Infectious SARS-CoV-1, SARS-CoV-2 or MERS-CoV particles in the supernatant of (A) as assessed by plaque-forming units assay. Panel C shows primary data.
(D) Viral E gene RNA levels in the supernatant of Calu-3 48 h post-infection with SARS-CoV-1 or 24 h post-infection with MERS-CoV. Cells were transiently transfected with control siRNA (CTRL) or siRNA targeting IFITM2 or IFITM3 as indicated. Cells were either treated with IFN-β (100 U/ml for SARS-CoV-1 or 50 U/ml for MERS-CoV) or left untreated.
(E) Infectious SARS-CoV-1 or MERS-CoV particles in the supernatant of (D) as assessed by plaque-forming units assay. Bars in all panels represent the mean of three independent experiments (±SEM); ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001. See also Figure S2.
IFITM2 and IFITM3 are highly homologous7 and it has been reported that both localize in early and late endosomes,31,32 which are major sites of SARS-CoV-1 and MERS-CoV entry.12,13,14 Thus, we also examined whether IFITM3 affects infection of SARS-CoV-1 and MERS-CoV. Silencing of both IFITM2 and IFITM3 impaired SARS-CoV-1 replication and infectious virus production in the absence of IFN by about one order of magnitude but had less than 2-fold effects on MERS-CoV (Figures 4D, 4E, and S2). To avoid saturating effects, we used a 5-fold lower concentration of IFN-β compared to the previous setting (Figures 4A–4C). Under these conditions of IFN-induced IFITM expression, siRNA silencing of both IFITM2 and IFITM3 reduced SARS-CoV-1 RNA production and infectious virus yields by ∼30- to 40-fold (Figures 4D, 4E, and S2). In comparison, replication of MERS-CoV was only diminished by 3- to 6-fold. Altogether, these results showed that endogenous expression of IFITMs is critical for efficient replication of hCoV-OC43, SARS-CoV-1 and SARS-CoV-2, but has little if any effect on hCoV-229E, hCoV-NL63 and MERS-CoV infection.
Potential interactions between hCoV spike proteins and IFITMs
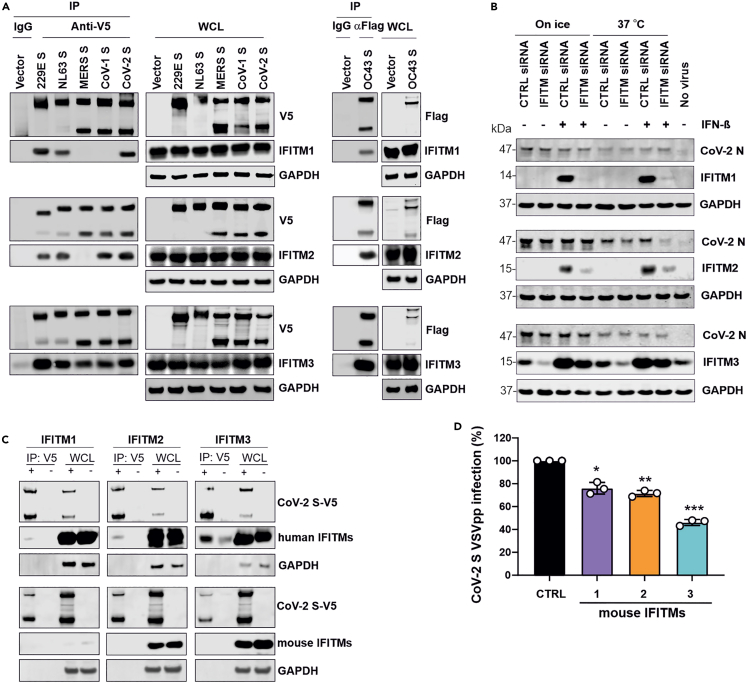
Our previous results from proximity ligation and mammalian-membrane two-hybrid assays as well as co-immunoprecipitation analyses, strongly suggested that IFITM-dependent SARS-CoV-2 infection requires specific interactions of IFITMs with the viral S protein.22 To examine this further, we performed comprehensive co-immunoprecipitation studies and found that the hCoV-229E,-NL63,-OC43 and SARS-CoV-2 S-proteins co-purified with all three IFITM proteins (Figure 5A). In comparison, the MERS-CoV S did not co-precipitate IFITM1 and IFITM2 and the SARS-CoV-1 S failed to pull down IFITM1. Notably, the S proteins of all CoVs exploiting IFITMs, i.e., hCoV-OC43 (IFITM3), SARS-CoV-1 (IFITM2 and IFITM3) and SARS-CoV-2 (IFITM2), interacted with the IFITM proteins that were most critical for efficient infection. To determine potential effects on virion binding to the cells, Calu-3 cells were transfected with IFITM-targeting or non-targeting siRNAs, treated with IFN-β or left untreated, and exposed to SARS-CoV-2 (MOI of 2.5) for 2 h on ice or at 37°C. Western blot analyses showed that different levels of IFITM expression had no significant effects on the amounts of cell-associated SARS-CoV-2 nucleocapsid (N) (Figure 5B). Altogether, the results suggest that interactions between CoV S proteins and IFITMs may be required but not sufficient for effective utilization of IFITMs as infection cofactors.
Figure 5.
Co-immunoprecipitation of IFITM proteins by CoV S proteins
(A) HEK293T cells were cotransfected with empty control vector or constructs expressing V5- or Flag-tagged 229E, NL63, OC43, MERS, CoV-1 and CoV-2 S proteins and IFITM1, IFITM2, or IFITM3, as indicated. 48h post-transfection, cell lysates were harvested for Co-IP assays. Similar results were obtained in two independent experiments.
(B) Calu-3 cells were transfected with either non-targeting siRNA or siRNA targeting IFITM1, IFITM2 or IFITM3 and treated with IFN-β (500U/ml) or left untreated. Cell were incubated on ice or at 37°C for 1 h, exposed to SARS-CoV-2 (MOI 2.5), and further incubated on ice or at 37°C for 2 h. Subsequently, cells were washed once by PBS and lysed for western blot analysis using the indicated antibodies. Shown is one representative example of three western blots performed.
(C) HEK293T cells were cotransfected with construct expressing V5-tagged CoV-2 S proteins and Flag-tagged mouse IFITMs. Whole cell lysates were harvested at 48h post-transfection for Co-IP analysis. Similar results were obtained in two independent experiments.
(D) Huh7 cells overexpressing the indicated mouse IFITMs were infected with VSVpp carrying the SARS-CoV-2 S protein. GFP+ cells were determined 24 h post-infection. Bars represent the mean of three independent experiments (±SEM). ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p > 0.001.
It has recently been reported that IFITM3 knockout (KO) mice demonstrate increased susceptibility to SARS-CoV-2 induced disease.33 A protective effect of mouse IFITM3 in vivo may seem at odds with a role of IFITMs as critical cofactors for efficient SARS-CoV-2 infection. To address this, we examined whether the broad inhibitory effect but not the specific interaction between the S protein in IFITMs may be conserved between humans and mice. We found that the SARS-CoV-2 S co-immunoprecipitated human but not mouse IFITM2 and IFITM3 proteins (Figure 5C). In comparison, overexpression of both human and mouse IFITMs inhibited S-mediated infection by VSVpp (Figures 2B and 5D). Thus, conservation of the broad inhibitory function and lack of specific interactions with the viral spike may explain protective effects of IFITM3 in mouse models.
Overexpression of IFITM proteins prevents ACE2 cell surface expression
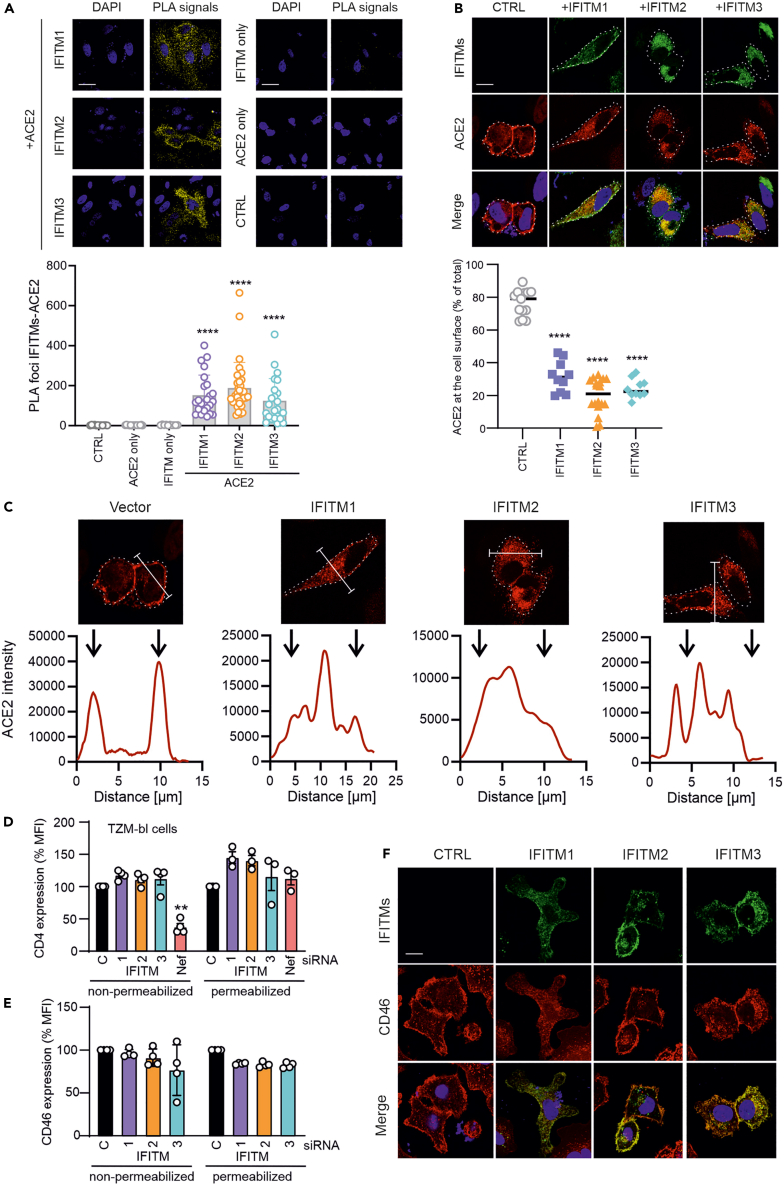
Increasing evidence supports that endogenously expressed IFITMs promote infection by SARS-CoVs, whereas overexpression inhibits it.20,21 To elucidate the mechanisms underlying these opposing effects, we analyzed whether IFITMs may interact with ACE2 and/or affect the subcellular localization of this primary receptor of SARS-CoV-1, SARS-CoV-2 and hCoV-NL63. To assess potential interactions, we co-transfected HeLa cells with constructs expressing ACE2 and the three IFITM proteins and performed proximity ligation assays (PLA). The results revealed high numbers of PLA foci for ACE2 and all three IFITMs in the cytoplasm rather than the cell surface (Figure 6A). These results showed that ACE2 and IFITM proteins are in close proximity and may interact in living cells. To examine potential effects on subcellular localization, we cotransfected HeLa with vectors expressing ACE2 and either IFITM1, IFITM2 or IFITM3 and analyzed them by confocal microscopy. As expected, ACE2 was mainly detected at the surface of HeLa cells transfected with the control vector (Figure 6B). In striking contrast, the proportion of ACE2 localized at the cell surface dropped from ∼80% to ∼20% on IFITM overexpression (Figures 6B and 6C). Similar results were obtained in A549-TMPRSS2-ACE2 transfected with a control vector or constructs expressing IFITM1, IFITM2 or IFITM3 (Figure S3A). Flow cytometric analyses of permeabilized and non-permeabilized cells confirmed that overexpression of IFITM1, IFITM2 or IFITM3 significantly reduced the levels of ACE2 exposed at the surface of Hela-ACE2 and A549-TMPRSS2-ACE2 cells but not the total expression levels (Figures S3B and S3C). The effect of IFITM overexpression was specific for ACE2 because it had no significant impact on cell surface expression of CD4 and CD46, the primary receptors of HIV and measles virus, respectively (Figures 6D–6F).
Figure 6.
IFITM overexpression inhibits ACE2 cell surface expression
(A) Proximity ligation assay between ACE2 and IFITM1, IFITM2 or IFITM3 in transfected HeLa cells. Scale bar, 20 μm. Bars represent the mean of two independent experiments (±SEM), ∗∗∗∗p < 0.0001.
(B) HeLa-ACE2 cells were transfected with IFITM1, IFITM2 or IFITM3 expression constructs or an empty control vector. Forty-eight hours post-transfection cells were stained with anti-ACE2 and anti-IFITMs antibodies and examined by confocal microscopy. Scale bar, 20 μm. ACE2 signal intensities at the cell surface and in the cytoplasm were quantified using ImageJ. ∗∗∗∗p < 0.0001.
(C) Intensity of ACE2 staining throughout the cell in the absence and presence of IFITM overexpression quantified using ImageJ.
(D and E) TZM-bl cells (D) or A549 stably expressing TMPRSS2 and ACE2 (E) were transfected with either IFITM1, IFITM2, or IFITM3 expression constructs. 48 h post-transfection, cells were permeabilized (left panels) or not (right panels), stained with anti-CD4 or anti-CD46 antibody respectively and analyzed by flow cytometry. Shown are mean fluorescence intensities (MFIs) measured in cells transfected with IFITM expression vectors relative to those that received the control construct (100%). Bars represent the mean of four independent experiments (±SEM), ∗∗, p < 0.01.
(F) A549 stably expressing TMPRSS2 and ACE2 cells were transfected with flag-tagged IFITM1, IFITM2 or IFITM3 expression constructs or an empty control vector. Forty-eight hours post-transfection cells were stained with anti-CD46 and anti-flag antibodies and examined by confocal microscopy. See also Figure S3.
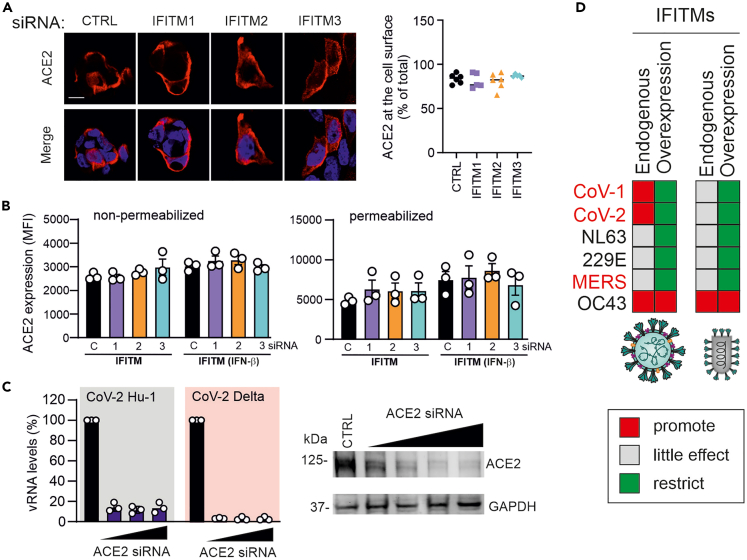
The above-mentioned results raised the possibility that silencing of endogenous IFITM expression may enhance ACE2 cell surface expression. Unlike IFITM overexpression, however, siRNA silencing of endogenous IFITM had no significant effect on ACE2 (Figure 7A). In agreement with published data,34 IFN-β moderately increased the levels of ACE2. Silencing of IFITMs, however, neither affected ACE2 expression nor the proportion accessible at the cell surface (Figure 7B). To challenge our hypothesis that IFITM overexpression inhibits SARS-CoV-2 by reducing ACE2 surface expression, we silenced expression of this receptor by siRNA treatment of Calu-3 cells. Reduced ACE2 expression strongly inhibited replication of the original SARS-CoV-2 Hu-1 strain and the Delta variant of concern in Calu-3 cells (Figure 7C). Thus, efficient ACE2 cell surface expression is critical for SARS-CoV-2 infection and impaired by overexpression of IFITM proteins. Taken together, our results demonstrate that artificially overexpressed IFITMs traps ACE2 in the cytoplasm, whereas endogenous IFITM expression does not affect ACE2 localization.
Figure 7.
ACE2 cell surface expression is not affected by silencing of endogenous IFITM expression and critical for SARS-CoV-2 infection
(A) Calu-3 cells were transfected twice with non-targeting control (NT) or with IFITM1, 2 or 3 siRNAs (with or without IFN-β treatment). Six hours after the second transfection, cells were stained with anti-ACE2 antibody and images acquired using the LSM system. ACE2 signal intensities at the cell surface and in the cytoplasm were quantified using ImageJ.
(B) Calu-3 cells were transfected twice with NT or IFITM1, 2 or 3 siRNAs as described in panel A, permeabilized or not, stained with anti-ACE2 antibody and analyzed by flow cytometry. Bar diagrams in panel B and C represent the mean of three independent experiments (±SEM).
(C) Calu-3 cells were treated with NT or ACE2 siRNAs, infected with the SARS-CoV-2 Hu-1 or Delta variants (MOI 0.05) and viral RNA production in the cell culture supernatants determined by qRT-PCR two days later.
(D) Schematic heatmap summarizing the effects of IFITMs on S-containing viral pseudo-particles and genuine hCoVs. Note that this overview is provisional and results may vary depending on the experimental conditions and cell types used.
Discussion
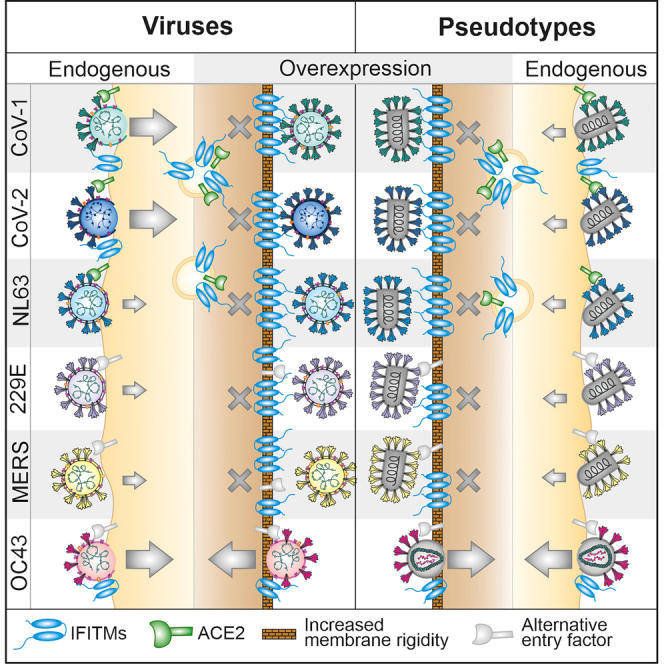
To elucidate the enigmatic role of IFITMs in human coronavirus infection and to clarify seemingly contradictory previous findings, we investigated the influence of transient and endogenous IFITM expression on infection by spike-containing viral pseudo-particles, as well as six of the seven genuine human coronaviruses. We show that endogenous expression of IFITMs boosts replication of SARS-CoV-1, SARS-CoV-2 and hCoV-OC43 but has little if any effect on hCoV-NL63, hCoV-229E and MERS-CoV infection (Figure 7D). In addition, we demonstrate that overexpression of IFITMs prevents cell surface expression of ACE2 and consequently inhibits infection by ACE2-tropic SARS-CoV-1, SARS-CoV-2 and hCoV-NL63. Our results explain the reported opposing effects of IFITMs and further establish them as critical cofactors for efficient replication of SARS-CoV-1 and SARS-CoV-2 in human lung cells.
Pseudo-typed viral particles have proven highly useful to examine the activity of neutralizing antibodies and other agents targeting the spike proteins of human coronaviruses including SARS-CoV-2. However, IFITMs are thought to exert their antiviral activity by affecting the composition and fluidity of cellular membranes instead of specifically engaging spike or other viral proteins.12,13,14 Previous studies reporting inhibitory effects of IFITMs used MLV-GFP pseudo-types carrying CoV-1 S,21 or lentiviral particles carrying S proteins of different CoVs including SARS-CoV-2 and MERS-CoV.19,20 Our present and earlier22 results obtained using S-containing HIV- and VSV-based pseudo-particles agree with these previous findings. However, shapes, lipid composition and curvatures of S-containing pseudo-virions differ drastically from genuine CoV particles and more closely resemble those of the carrier virus. In addition to the S protein, infectious CoV particles also contain the membrane (M) and envelope (E) proteins as essential components of their lipid bilayer.35 The M protein is highly abundant in CoV particles and may bind to target cells using heparan sulfate proteoglycans as initial attachment factors.36 Thus, although single-round pseudo-viruses may reflect key aspects of S-mediated virus attachment and its inhibition, it is conceivable that they do not represent faithful models for attachment and fusion of genuine hCoV particles. Consequently, our data indicate that viral pseudo-particles carrying SARS-CoV-1 or SARS-CoV-2 S-proteins do not reproduce the ability of the native viruses to hijack IFITMs for efficient infection. Potential reasons for this are that the pseudo-virions may be particularly susceptible to the membrane modulating effects of IFITMs and/or that the ability of SARS-CoVs to exploit IFITMs for infection requires specific features of the genuine viral particles.
The broad antiviral effect of IFITM as a modulator of membrane rigidity does not explain why endogenous and artificially overexpressed IFITMs have opposing effects on genuine SARS-CoV-2 infection.19,22 Here, we demonstrate that no less than three of the five human β coronaviruses, hCoV-OC43, SARS-CoV-1 and SARS-CoV-2, require IFITMs for efficient replication. SARS-CoV-1 and SARS-CoV-2 are closely related and both utilize ACE2 as major receptor, just like hCoV-NL63.37,38,39 We discovered that artificial overexpression of IFITM1, 2 and 3 prevents ACE2 expression at the cell surface. Our ongoing studies suggest that lower expression levels at least in part explain why endogenously expressed IFITMs do not compromise ACE2 cell surface expression. Because ACE2 is essential for infection by SARS-CoV-1, SARS-CoV-2 and hCoV-NL63, this effect provides a plausible explanation for previously reported inhibitory activities of IFITM overexpression. Our results agree with the recent finding that ACE2 is required for inhibitory effects of IFITM3 on SARS-CoV-2 S-mediated infection.40 It also explains the diverse effects of IFITM overexpression on common cold coronaviruses: inhibition of hCoV-NL63 is particularly effective because this ACE2-tropic virus should be affected by both changes in membrane fluidity and down-modulation of ACE2. In contrast, IFITM3 overexpression enhances hCoV-OC43 infection because this virus exploits IFITM3 for infection and does not use ACE2 as entry receptor. Thus, the entry receptors of OC43 remain available even under conditions of IFITM overexpression allowing robust enhancement by IFITM3. Finally, hCoV-229E exploits neither ACE2 nor IFITMs for infection and showed moderate sensitivity to inhibition by IFITM overexpression that is most likely because of IFITM-mediated changes in cellular membrane rigidity.
Although knock-down of endogenous IFITM expression had little, if any, effect on hCoV-229E, hCoV-NL63 and MERS-CoV it strongly reduced replication of SARS-CoV-1, SARS-CoV-2 and hCoV-OC43 even in the absence of IFN treatment. This shows that low basal levels of IFITM expression are sufficient to boost infection by several hCoVs, whereas marked inhibition of genuine viruses requires overexpression. The exact mechanisms underlying IFITM-dependent enhancement of infection remain to be determined. However, results of co-immunopreciptation and PLA assays, together with the inhibitory effect of anti-IFITM2 antibodies22,41 suggest that SARS-CoV-2 S proteins may interact with the N-terminal region of IFITMs to promote virion attachment or entry. We found that IFITM expression did not increase the levels of cell-associated SARS-CoV-2 nucleocapsid (N) protein after brief (2 h) exposure of Calu-3 cells to viral particles (Figure 5B). However, our previous studies showed that depletion of IFITM2 significantly reduces the intracellular levels of viral RNA already 6 h after virus exposure.22 Thus, endogenous IFITM2 expression promotes an early step of SARS-CoV-2 infection but does not seem to have a major impact on the overall levels of virus attachment. Our finding that IFITMs are in close proximity to spike and ACE2 raises the possibility that both may cooperate to promote SARS-CoV entry, although further studies are required to challenge this hypothesis.
SARS-CoV-1 and SARS-CoV-2 are highly similar to one another but only distantly related to hCoV-OC43. The finding that all three hCoVs hijack IFITMs for efficient infection, suggests that utilization of IFITMs as entry cofactors may be more common among coronaviruses than previously anticipated and has possibly been independently acquired several times during their evolution. In line with this possibility the underlying mechanisms appear to differ. The OC43 S exploits IFITM3 irrespectively of the carrier particle.18 In contrast, IFITMs specifically enhance infection of genuine SARS-CoVs but not of the corresponding S-containing HIV- or VSV-based particles.22,23 It has been shown that the C-terminal domain of IFITM3 is essential for enhancement of hCoV-OC43 infection,18 whereas the N-proximal region of IFITM2 seems critical for enhancement of SARS-CoV-2 infection. Further analyses of the interactions between IFITMs, S proteins, and ACE2 at sites of viral fusion are required to elucidate how SARS-CoVs can use these otherwise antiviral proteins for efficient entry into their host cells.
It has been reported that knock-out of IFITM3 in transgenic mice expressing human ACE2 is associated with more severe disease and early death compared to control mice.33 A protective effect of IFITM3 against severe disease in SARS-CoV-2 infected mice may seem at odds with our finding that IFITMs are important cofactors of SARS-CoV-2 infection. However, as antiviral restriction factors, IFITMs are under high pressure for positive selection and are thus highly variable.42 We found that mouse IFITMs do not interact with the SARS-CoV-2 S protein in co-immunoprecipitation assays but display antiviral activity. This agrees with findings that overexpression of mouse IFITMs inhibits and silencing of endogenous mouse IFITM3 promotes SARS-CoV-2 infection.19 Altogether, these results suggest that the broad antiviral activity of IFITMs but not the hijacking by SARS-CoV-2 S proteins is conserved between humans and mice.
IFITMs are expressed in human lung, gut, heart and brain cells and strongly induced in SARS-CoV-2 infected individuals.22,43,44 High levels of expression at the primary targets for viral transmission and dissemination make IFITMs particularly suitable to be used as entry cofactors. We and others have shown that IFITM-targeting antibodies may inhibit SARS-CoV-2 infection.22,41 In addition, recent data identified IFITMs as SARS-CoV-2 dependency factors.45 Thus, increasing evidence suggests that IFITMs may be suitable targets for therapeutic approaches against SARS coronaviruses and further studies on their role in viral dissemination and pathogenesis are highly warranted.
Limitations of the study
We demonstrate that three human coronaviruses including the two responsible for severe respiratory disease utilize IFITMs for efficient replication. Our results further demonstrate that down-modulation of cell surface ACE2 explains why artificial IFITM overexpression inhibits ACE2-tropic hCoVs. However, the exact mechanism(s) underlying the enhancing effects of IFITMs on SARS-CoV-1, SARS-CoV-2 and hCoV-OC43 infection remain to be defined. It will also be interesting to determine whether isoforms of IFITM2 that differ in their antiretroviral activity,46 also differ in their ability to promote SARS-CoV-1 and SARS-CoV-2 infection. In addition, further studies are required to clarify to relevance of IFITMs for transmission, tissue tropism and pathogenesis of SARS-CoVs in vivo. Finally, our results suggest that IFITMs may directly interact with the spike proteins of human coronaviruses and perhaps also ACE2. Detailed functional and structural characterization of these interactions might allow us to develop antivirals targeting IFITMs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| IRDye 680RD Goat anti-Rabbit IgG (H + L) | LI-COR | Cat926-68071 |
| IRDye 800CW Goat anti-Mouse IgG (H + L) | LI-COR | Cat# 926-32210 |
| IRDye 800RD Goat anti-Rat IgG (H + L) | LI-COR | Cat# 925-32219 |
| HCoV-229E Nucleocapsid Antibody | SinoBiological | Cat# 40640-T62 |
| Anti-Coronavirus NL63 | Ingenasa | M.30.HCo.I2D4 |
| Anti-Coronavirus Antibody, OC-43 strain | Millipore | Cat# MAB9012 |
| Purified anti-GAPDH Antibody | Biolegend | Cat# 607902 |
| Rabbit polyclonal anti-IFITM1 | Cell signaling | Cat# 13126 S |
| Rabbit polyclonal anti-IFITM2 | Cell signaling | Cat# 13530S |
| Rabbit polyclonal anti-IFITM3 | Cell signaling | Cat# 59212S |
| V5-Tag (E9H8O) Mouse mAb | Cell signaling | Cat# 80076 |
| SARS-CoV-2 (2019-nCoV) Nucleocapsid Antibody | SinoBiological | Cat# 40588-MM124 |
| Rabbit polyclonal anti-ACE2 | Abcam | Cat# ab15348 |
| Mouse monoclonal anti-flag | Sigma-Aldrich | Cat# F1804 |
| Fixable Viability Dye eFluor 780 | Thermo Fisher | Cat# 65-0865-14 |
| Rabbit monoclonal Alexa Fluor® 647 anti-CD46 | Abcam | Cat# 275208 |
| Rat Monoclonal PerCP-Cy5.5 anti-CD4 | Biolegend | Cat# 317428 |
| Bacterial and virus strains | ||
| VSVΔG(GFP)∗VSV-G | Klaus Conzelmann | N/A |
| hCoV-229E | ATCC | Cat# VR-740TM |
| hCoV-NL63 | Lia van der Hoek | N/A |
| hCoV-OC43 | ATCC | Cat# CR-1558TM |
| Renilla hCoV-229E | Volker Thiel | N/A |
| BetaCoV/Netherlands/01/NL/2020 | European Virus Archive | Cat# 010V 03903 |
| VSVΔG(GFP)∗VSV-G | Klaus Conzelmann | N/A |
| BetaCoV/Netherlands/01/NL/2020 | European Virus Archive | Cat# 010V 03903 |
| BetaCoV/B.1.617.2/Delta | Florian Schmidt (University of Bonn) | N/A |
| BetaCoV/Munich/BavPat1/2020 | GISAID; Wölfel et al., 202047 | EPI_ISL_406862 |
| SARS-CoV (rSCV, Frankfurt-1 virus isolate) | Pfefferle et al., 200929 | Genbank accession no. AY310120 |
| rMERS-CoV, MERS-CoV/EMC2012 | Muth et al., 201748 | Genbank accession no. NC_019843.3 |
| Chemicals, peptides, and recombinant proteins | ||
| L-Glutamine | Pan Biotech | Cat# P04-80100 |
| DAPI | Sigma-Aldrich | Cat# D9542-1MG; CAS: 28718-90-3 |
| Dulbecco’s Modified Eagle Medium (DMEM) | Gibco | Cat# 41965039 |
| Dulbecco’s Phosphate Buffered Saline (PBS) | Thermo Fisher | Cat# 14190094 |
| Minimum Essential Medium Eagle (MEM) | Sigma-Aldrich | Cat# M4655 |
| Opti-MEM | Gibco | Cat# M3942 |
| Roswell Park Memorial Institute (RPMI) | Gibco | Cat# 22400089 |
| Penicillin-Streptomycin | Thermo Fisher | Cat# 15140122 |
| non-essential amino acids | Sigma-Aldrich | Cat# M7145 |
| Sodium pyruvate | Sigma-Aldrich | Cat# S8636 |
| FCS | Gibco | Cat# 10270106 |
| Trypsin EDTA | Gibco | Cat# T3924 |
| Versene | Thermo Fisher | Cat# 15040066 |
| TMB peroxidase substrate (Sure blue) | Medac | Cat# 52-00-04 |
| BD Cytofix/Cytoperm™ Fixation/Permeabilization Solution Kit | BD Biosciences | Cat# 554714 |
| Lipofectamine RNAiMAX | ThermoFisher | Cat# 13778075 |
| SuperScript™ III Platinum™ One-Step qRT-PCR Kit | Invitrogen™ | Cat# 11732088 |
| Renilla Luciferase Assay kit | Promega | Cat# E2810 |
| Quick RNA Miniprep Kit | ZYMO research | Cat# R1055 |
| QIAamp Viral RNA Kits | QIAGEN | Cat# 52906 |
| Luciferase Assay Kit | Promega | Cat# E1500 |
| Mowiol 4-88 | ROTH | Cat# 0713.1 |
| Blocker Casein in PBS | Thermo Fisher | Cat# 37528 |
| TaqMan Fast Virus 1-Step Master Mix | ThermoFisher | Cat# 4444436 |
| Tween 20 | Sigma-Aldrich | Cat# P7949500ML |
| Triton X-100 | Sigma-Aldrich | Cat#T9284 100ML |
| GAPD (GAPDH) Endogenous Control | ThermoFisher | Cat# 4352338E |
| β-Mercaptoethanol | Sigma-Aldrich | Cat# 444203 |
| Immobilon-FL PVDF membranes | Merck Millipore | Cat# IPVH00010 |
| Paraformaldehyde | Merck, Darmstadt | Cat# 104005 |
| TransIT-LT1 | Mirus | Cat# MIR2300 |
| NuPAGE 4±12% Bis-Tris Gels | ThermoFisher | Cat# NP0321BOX |
| Synthetic SARS-CoV-2-RNA | Twist Bioscience | Cat# 102019 |
| RNAiMAX | ThermoFisher | Cat# LMRNA015 |
| Recombinant IFN-α2 | pbl assay science | Cat# 11101-1 |
| Recombinant IFN-β | R&D systems | Cat# 8499-IF-010 |
| Recombinant IFN-γ | Sigma-Aldrich | Cat# I3265 |
| Recombinant IFN- λ3 | CHO | Cat# 5259-IL-025 |
| Cytochalasin D | ThermoFisher | Cat# PHZ1063 |
| Agarose | Sigma-Aldrich | Cat# A9539 |
| Experimental models: Cell lines | ||
| Human: Calu-3 | ATCC or M. Frick | Cat# HTB-55 or N/A |
| Human: Caco2 | Holger Barth | |
| Human: Huh7.5 | Ralf Bartenschlager | |
| Human: Huh7 | Anna-Laura Kretz | |
| LLCMK2 | by Lia van der Hoek | |
| HCT8 | ATCC | CCL-244TM |
| Human: HEK293Ts | ATCC | Cat# CRL-3216 |
| Human: HeLa | ATCC | Cat# CRM-CCL-2 |
| African Green Monkey: Vero B4 | ATCC | Cat# DSMZ-AC33 |
| African Green Monkey: Vero E6 | ATCC | Cat# DSMZ-AC33 |
| Human: Hela-ACE2 | This paper | Cat# CRL-1586 |
| Human: A549 | Hoffmann et al., 202037 | |
| Human: A549-ACE2 | Hoffmann et al., 202037 | |
| Human: A549-TMPRSS2 | Hoffmann et al., 202037 | |
| Human: A549-TMPRSS2-ACE2 | Hoffmann et al., 202037 | |
| Human: TZM-bl | NIH | Cat# ARP-8129 |
| Oligonucleotides | ||
| 229E Forward: TACTCGAGGTTCCGTCTC GT | Biomers | N/A |
| 229E Reverse: CAAGTGTCACACGGTTGCAG | Biomers | N/A |
| 229E probe: AGC AAA GTT CTG TCT TTG TGG AAA CCA | Biomers | N/A |
| NL63 Forward: AAACCTCGTTGGAAGCGTGT | Biomers | N/A |
| NL63 Reverse: CTGTGGAAAACCTTTGGCATC | Biomers | N/A |
| NL63 Probe: ATGTTATTCAGTGCTTTGGTCCTCGTGAT | Biomers | N/A |
| OC43 Forward: AGCAACCAGGCTGATGTC AATACC | Biomers | N/A |
| OC43 Reverse: AGCAGACCTTCCTGAGCCTTCAAT | Biomers | N/A |
| OC43 Probe: TGACATTGTCGATCGGGACCCAAGTA | Biomers | N/A |
| HKU-NF forward: TAATCAGACAAGGAACTGATTA | Biomers | N/A |
| HKU-N reverse: CGAAGGTGTGACTTCCAT G | Biomers | N/A |
| HKU-N Probe: FAM-GCAAATTGTGCAATTTGCGG-TAMRA | Biomers | N/A |
| SARS-CoV-1 forward (EMC-upE-F): GCA ACG CGC GAT TCA GTT |
Biomers | N/A |
| SARS-CoV-1 reverse (EMC-upE-R): GCC TCT ACA CGG GAC CCA TA |
Biomers | N/A |
| SARS-CoV-1 probe (EMC-upE-P): FAM-CTC TTCACATAATCGCCCCGAGCTCG-TAMRA | Biomers | N/A |
| SARS-CoV-2 forward (HKU-NF): TAA TCA GAC AAG GAA CTG ATT A | Biomers | N/A |
| SARS-CoV-2 reverse (HKU-NR): CGA AGG TGT GAC TTC CAT G | Biomers | N/A |
| SARS-CoV-2 probe (HKU-NP): FAM-GCA AAT TGT GCA ATT TGC GG-TAMRA | Biomers | N/A |
| MERS-CoV forward (E-Sarbeco-F): ACA GGT ACG TTA ATA GTT AAT AGC GT | Biomers | N/A |
| MERS-CoV reverse (E-Sarbeco-R): ATA TTG CAG CAG TAC GCA CAC A | Biomers | N/A |
| MERS-CoV probe (E-Sarbeco-P): FAM-ACA CTA GCC ATC CTT ACT GCG CTT CG-BBQ | Biomers | N/A |
| Human ACTB (Beta Actin) Endogenous Control | Applied Biosystems | Cat# 4310881E |
| CTRL siRNA-01: UAAGGCUAUGAAGAGAUAC | This paper | D-001206-14-20 |
| CTRL siRNA-02: AUGUAUUGGCCUGUAUUAG | This paper | D-001206-14-21 |
| CTRL siRNA-03: AUGAACGUGAAUUGCUCAA | This paper | D-001206-14-22 |
| CTRL siRNA-04: UGGUUUACAUGUCGACUAA | This paper | D-001206-14-23 |
| siRNA IFITM1-01: GUGACAGUCUACCAUAUUA | Dharmacon | D-019543-01 |
| siRNA IFITM1-02: CUUCCAAGGUCCACCGUGA | Dharmacon | D-019543-02 |
| siRNA IFITM1-03: CCUCAUGACCAUUGGAUUC | Dharmacon | D-019543-03 |
| siRNA IFITM1-04: GUUACAGAUAAUACAGGAA | Dharmacon | D-019543-04 |
| siRNA IFITM2-01: CAAACCUUCUCUCCUGUCA | Dharmacon | D-020103-01 |
| siRNA IFITM2-02: UCAAGGAGGAGCAGGAAGU | Dharmacon | D-020103-02 |
| siRNA IFITM2-03: UCGUCCAGGCCCAGCGAUA | Dharmacon | D-020103-03 |
| siRNA IFITM2-04: UGGUCUGGUCCCUGUUCAA | Dharmacon | D-020103-18 |
| siRNA IFITM3-01: ACGUGUUUCUGGUGCUAAA | Dharmacon | D-014116-13 |
| siRNA IFITM3-02: AUGGAUAGAUCAGGAGGCA | Dharmacon | D-014116-14 |
| siRNA IFITM3-03: UGCUGAUCUUCCAGGCCUA | Dharmacon | D-014116-15 |
| siRNA IFITM3-04: UCGUCAUCCCAGUGCUGAU | Dharmacon | D-014116-16 |
| siRNA ACE2-01: GAACAUACCUUUGAAGAGA | Dharmacon | D-005755-01 |
| siRNA ACE2-02: GCACAAAGGUGACAAUGGA | Dharmacon | D-005755-02 |
| siRNA ACE2-03: GAAGGUGGAUGGUCUUUAA | Dharmacon | D-005755-03 |
| siRNA ACE2-04: GAGGGAGACUAUGAAGUA | Dharmacon | D-005755-04 |
| Recombinant DNA | ||
| pCG_hCoV-229E-spike C-V5-tag_BFP | Lawrenz et al., 202249 | |
| pCG_hCoV-NL63-spike C-V5-tag_BFP | Lawrenz et al., 202249 | |
| pCMV3-hCoV-OC43-spike-Flag | Sinobiological | VG40607-CF |
| pCMV3-hCoV-HKU1-spike | Sinobiological | VG40021-UT |
| pCG_MERS-CoV S (humanized) C-V5 IRES GFP | Michael Schindler | |
| pCG_SARS-CoV-1 S (hum) C-V5 IRES GFP | Michael Schindler | |
| pCG_SARS-CoV-2-Spike-V5_IRES_eGFP (humanized) | Prelli Bozzo et al., 202122 | |
| pCG_IFITM1 | Prelli Bozzo et al., 202122 | N/A |
| pCG_IFITM2 | Prelli Bozzo et al., 202122 | N/A |
| pCG_IFITM3 | Prelli Bozzo et al., 202122 | N/A |
| Mouse Ifitm1(NM_026820) ORF Clone | Genescript | OMU15399D |
| Mouse Ifitm2(NM_030694) ORF Clone | Genescript | OMU08371D |
| Mouse Ifitm3(NM_025378) ORF Clone | Genescript | OMU18001D |
| pCG_IFITM1-IRES_eGFP | Prelli Bozzo et al., 202122 | N/A |
| pCG_IFITM2-IRES_eGFP | Prelli Bozzo et al., 202122 | N/A |
| pCG_IFITM3-IRES_BFP | Prelli Bozzo et al., 202122 | N/A |
| pTwist_EF1a_3Xflag_IFITM1 | This paper | N/A |
| pTwist_EF1a_3Xflag_IFITM2 | This paper | N/A |
| pTwist_EF1a_3Xflag_IFITM3 | This paper | N/A |
| pCDNA3.1_hACE2 | This paper | N/A |
| pCDNA3.1_hCD4 | This paper | N/A |
| pCG_Nef NL4-3_IRES GFP | This paper | N/A |
| pCG_SARS-CoV2-Spike-IRES_eGFP | Stefan Pöhlmann37 | N/A |
| Software and algorithms | ||
| Corel DRAW 2017 | Corel Corporation | https://www.coreldraw.com/ |
| GraphPad Prism Version 9 | GraphPad Software, Inc. | https://www.graphpad.com |
| LI-COR Image Studio Lite Version 5.0 | LI-COR | www.licor.com/ |
| Fiji (ImageJ) version 1.8 | Java | www.imagej.nih.gov/ij |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact Frank Kirchhoff (frank.kirchhoff@uni-ulm.de).
Materials availability
All unique reagents generated in this study are listed in the key resources table and available from the lead contact.
Experimental model and subject details
Cell culture
All cells were cultured at 37°C in a 5% CO2 atmosphere. HEK293T cells (Human embryonic kidney 293T cells; ATCC) were cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 100 μg/ml streptomycin, 2 mM L-glutamine and 10% heat-inactivated fetal calf serum (FCS). A549 (adenocarcinomic human alveolar basal epithelial) parental cells were maintained in DMEM supplemented with 10% FCS, L-glutamine (2 mM), streptomycin (100 μg/ml), penicillin (100 U/ml), NEAA supplement (Non-essential amino acids (1 mM)) and sodium pyruvate (1 mM). A549 cells stably expressing ACE2, TMPRSS2 or both were maintained in the same culture medium as described for A549 parental cells, with additional Puromycin (2 μg/mL), Blasticidin (4 μg/mL) or both. Huh-7 cells and Huh7.5 cells were grown in DMEM supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, 2 mM L-glutamine, NEAA (1mM) and 10% FCS. HCT-8 (human ileocecal adenocarcinoma cells) cells were maintained in RPMI 1640 Medium which was supplemented with 100 units/ml penicillin, 100μg/ml streptomycin and 10% FCS. LLC-MK2 cells were maintained in DMEM supplemented with non-essential amino acids solution, 100 μg/ml streptomycin, 0.2 mM L-glutamine and 8% FCS. Caco-2 (human epithelial colorectal adenocarcinoma) cells were maintained in DMEM containing 20% FCS, glutamine (2 mM), streptomycin (100 μg/ml) and penicillin (100 U/ml), NEAA supplement (Non-essential amino acids (1 mM)), sodium pyruvate (1 mM). Calu-3 (human epithelial lung adenocarcinoma) cells were cultured in Minimum Essential Medium Eagle (MEM) supplemented with 10% FCS (during viral infection) or 20% (during all other times), penicillin (100 U/ml), streptomycin (100 μg/ml), sodium pyruvate (1 mM), and NEAA supplement (1 mM). Hela parental and Hela stably expressing ACE2 and TZM-bl cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FCS), L-glutamine (2 mM), streptomycin (100 μg/ml) and penicillin (100 U/ml).
Method details
Phylogenetic analysis
Reference nucleotide sequences of full-length human coronavirus genomes were obtained from the NCBI database (NC_005831.2; NC_006213.1; NC_006577.2; NC_002645.1; NC_045512.2; NC_004718.3; NC_019843.3), aligned using the NGPhylogeny.fr with MAFFT alignment (https://ngphylogeny.fr/).50,51 Phylogenetic tree was visualized using iTOL (https://itol.embl.de/).52
Expression constructs
Expression plasmids encoding spike proteins of hCoV-229E and hCoV-NL63, pCG_hCoV-229E-spike C-V5-tag_BFP and pCG_hCoV-NL63-spike C-V5-tag_BFP, were cloned as described.49 Expression plasmids encoding for IFITM1, IFITM2 and IFITM3 (pCG_IFITM1, pCG_IFITM2, pCG-IFITM3, pCG_IFITM1-IRES_eGFP, pCG_IFITM2-IRES_eGFP and pCG_IFITM3-IRES_BFP) were cloned as described.22 Expression plasmids encoding for flag-tagged IFITM1, IFITM2 and IFITM3 (pTwist_EF1a_ 3Xflag_IFITM1, pTwist_EF1a_3Xflag_IFITM2 and pTwist_EF1a_3Xflag_IFITM3) were PCR amplified and subcloned in pTwist based backbones using flanking restriction sites SpeI and NheI. The ORF of ACE2 was extracted with XbaI and MluI and then inserted into the XbaI-MluI site of pCG.
Pseudo-particle stock production
Vesicular stomatitis virus (VSV) based pseudoparticles wereproduced by transfection of HEK293T cells with spike expression plasmids and subsequent infection with VSV-glycoprotein (G) deleted but complemented (∗) VSV particles at MOI 3. Pseudotyped particles were harvested at 18 hours after infection. Residual particles carrying VSV-G were blocked by adding 10% (v/v) of I1 Hybridoma supernatant (I1, mouse hybridoma supernatant from CRL-2700; ATCC). To produce lentiviral particles containing hCoV-OC43 or hCoV-HKU1 spikes, HEK293T cells were transfected with spike expression construct together with a pNL1_HIV-1_NL4-3-Δenv-fluc back-bone. 48h after transfection, supernatants were harvested and stored at −80°C.
Virus stock production
For hCoV-229E propagation, Huh-7 cells were inoculated with a MOI of 0.1 in Dulbecco modified Eagle medium supplemented with 2% FCS. Cells were incubated at 33°C, and washed with phosphate-buffered saline and further cultured in fresh medium at 1 day post-infection. On day 4, supernatant was harvested, aliquoted, and stored at −80°C. HCoV-NL63 was propagated as described for hCoV-229E but using LLC-MK2 cells. HCoV-OC43 was propagated as described for hCoV-229E but using HCT-8 cells and harvesting viral stocks on day 6 and 8. BetaCoV/Netherlands/01/NL/2020 or B.1.617.2 (Delta) were propagated on Vero E6 infected at an MOI of 0.003 in serum-free medium as previously described.53 Briefly, the cells were inoculated for 2hat 37°C before the inoculum was removed. The supernatant was harvested 48 h post-infection upon visible cytopathic effect (CPE). To remove the debris, the supernatants were centrifuged for 5minat 1,000 × g, then aliquoted and stored at −80°C. Infectious virus titre was determined as plaque forming units (PFU). N gene RNA copies were determined by RT-qPCR. BetaCoV/Munich/BavPat1/2020 or rSCV were propagated on Vero E6, rMERS-CoV on Vero B4 cells infected with low passage virus stock solution (approximately 1,000,000 PFU per ml) in serum-free medium. Three days post-inoculation, supernatant was harvested upon visible cytopathic effect (CPE). To remove the debris, the supernatants were centrifuged for 5minat 1,000 × g and virus particles were purified from cytokines and concentrated using Vivaspin 20 (Sartorius, filtration units with a size exclusion of 100 kDa) according to the manufacturer’s instructions. Virus concentrate was resuspended in 2–3 ml PBS, diluted 1:2 in virus preservation medium (0.5% gelatine in OptiPRO serumfree medium) and stored at -80°C. Infectious virus titre was determined as plaque forming units (PFU) and viral RNA concentration was quantified by RT-qPCR. All stocks were sequenced by next generation sequencing methods and the absence of additional mutations was confirmed to occur in less than 20% of the virus-specific reads.
Renilla luciferase assay
To determine replication level of Renilla hCoV-229E, the supernatant of infected Huh7 cells was removed 24h postinfection and the cells lysed in 200μl of Renilla luciferase lysis buffer. Renilla luciferase of hCoV-229E was determined by Renilla Luciferase Assay Kit (Promega) according to the manufacturer’s instructions on an Orion microplate luminometer (Berthold).
TCID50 assay
To determine the infectious titer of hCoV-229E, 10,000 Huh-7 cells were seeded 1 day before infection in a 96-well plate. The following day, cells were inoculated with a 10-fold serial dilution of the respective virus stock. 4 days after infection, cytopathic effects were observed by light microscopy and the tissue culture infectious dose 50 (TCID50) was calculated according to Reed-Münch. For determining the TCID50 of hCoV-NL63 and hCoV-OC43 virus stocks, LLC-MK2 cells or HCT-8 cells were used and treated as described for hCoV-229E.
qRT-PCR
Total RNA from cell lysates or viral RNA from supernatant of seasonal coronaviruses-infected cells was isolated using Quick RNA Miniprep Kit (ZYMO research) or QIAamp Viral RNA Kits (QIAGEN) according to the manufacturer’s instructions. qRT-PCR was performed according to manufacturer’s instructions using SuperScript III Platinum One-11Step qRT-PCR Kit (Invitrogen) and OneStepPlus Real-Time PCR System. Primers and probes were ordered from Biomers and Applied Biosystems. N (nucleoprotein) RNA levels were determined in supernatants or cells collected from SARS-CoV-1, SARS-CoV-2 or MERS-CoV infected Calu-3 cells 48 h post-infection. Total RNA was isolated using the Viral RNA Mini Kit (Qiagen) according to the manufacturer’s instructions. RT-qPCR was performed according to the manufacturer’s instructions using TaqMan Fast Virus 1-Step Master Mix (Thermo Fisher) and a OneStepPlus Real-Time PCR System (96-well format, fast mode). Primers were purchased from Biomers and dissolved in RNAse free water. Synthetic SARS-CoV-2-RNA (Twist Bioscience) or linearized plasmids encoding N gene fragments (for ccCoVs) were used as a quantitative standard to obtain viral copy numbers. All reactions were run in duplicates.
Overexpression of IFITMs in human hepatocytes
24h after seeding, Huh7 cells were transfected with empty vector, pCG_IFITM1, pCG_IFITM2 or pCG_IFITM3 plasmids expressing corresponding IFITM proteins using TransIT-LT1 transfection reagent according to the manufacturer’s instructions. 3μl of TransIT-LT1 was used for per μg of plasmid transfection.
Plaque-forming units assay
The plaque-forming unit (PFU) assay was performed as previously described.53 SARS-CoV-1, SARS-CoV-2 and MERS-CoV stocks were serially diluted and confluent monolayers of Vero E6 or Vero B4 (for MERS-CoV) cells infected. After incubation for 1 to 3hat 37°C with shaking every 15 to 30 min. The cells were overlaid with 1.5 ml of 0.8% Avicel RC-581 (FMC) in medium and incubated for 3 days. Cells were fixed with 4% paraformaldehyde (PFA) at room temperature for 45 min. After the cells were washed with PBS once 0.5 ml of staining solution (0.5% crystal violet and 0.1% triton in water). After 20 min incubation at room temperature, the staining solution was removed using water, virus-induced plaque formation quantified, and PFU per ml calculated.
Co-immunoprecipitation
HEK293Ts were transfected with 1μg of spike proteins and 1 μg of pCG IFITM1, IFITM2, or IFITM3 or mouse IFITMs. 48h later, samples were collected and lysed in Co-IP lysis buffer (50 mM, Tris pH8, 150 mM NaCl, 1% NP40, protease inhibitor). The day after V5 antibodies or IgG isotype incubated with Pierce Protein A/G Magnetic beads (88802) (3μg of primary antibody per 7 μl of beads per sample), 200μg of whole cell lysates were added and incubated at 4 degrees. 4h after incubation, beads were washed 5 times with Co-IP lysis buffer and then boiled in 10% beta-Mercaptoethanol for 10minat 95 degrees.
IFITM1, 2 and 3 knock-down
24h after seeding, Huh7 cells were transfected with 45nM of either non-targeting siRNA or siRNAs targeting specific IFITMs using Lipofectamine RNAiMAX (Thermo Fisher) according to the manufacturer’s instructions. 24 h and 96 h post-seeding, Calu-3 cells were transfected with 45 nM twice (Calu-3) of either non-targeting siRNA or IFITM1, IFITM2 or IFITM3 specific siRNA using Lipofectamine RNAiMAX (Thermo Fisher) according to the manufacturer’s instructions. Prior to transfection, the medium was changed. 14 h posttransfection, the medium was replaced with 1 ml MEM supplemented with 500 U/ml IFN-β 7 h posttransfection, the Calu-3 cells were infected with SARS-CoV-2, SARS-CoV-1 or MERS-CoV (MOI 0.05) and 7 to 9 h later, supernatant was removed, and 1 ml fresh medium was added. 24h (MERS) or 48h post-infection cells and supernatants were harvested for Western blot and qPCR analysis respectively.
ACE2 knock-down
24 h and 96 h after seeding, Calu-3 cells were transfected twice with 45, 22.5, 11.25 or 5nM of either non-targeting or ACE2 specific siRNA using Lipofectamine RNAiMAX (Thermo Fisher) according to the manufacturer’s instructions. 14 h posttransfection, medium was replaced with fresh medium supplemented with 1000 U/ml IFN-β in the indicated conditions. 7 h after the second transfection, Calu-3 cells were infected with SARS-CoV-2 with an MOI of 0.05 and 2.5 respectively. 6 h later, the inoculum was removed, cells were washed once with PBS and supplemented with fresh media. 48 h post-infection, cells and supernatants were harvested for qRT-PCR analysis.
MTT assay
A549 stably expressing TMPRSS2 and ACE2 were seeded in a 96 well plate. The next day, cells were treated with 50μM of either Cytochalasin D for one hour. At the indicated timepoints, 10μl of MTT solution was added to the cells culture (1:10 diluted with PBS). 3h later, 200μl of DMSO and EtOH mixture (1:1) was added and the plates were measured at the luminometer (absorbance of 493 nm with the baseline correction at 650 nm).
Whole cell lysates
To determine expression of cellular and viral proteins, cells were washed in PBS and subsequently lysed in Western blot lysis buffer (150 mM NaCl, 50 mM HEPES, 5 mM EDTA, pH 7.4) supplemented with protease inhibitor as previously described.54 After 5 min of incubation on ice, the samples were centrifuged (4°C, 20 min, 14.000 rpm) to remove cell debris. The supernatant was transferred to a fresh tube, the protein concentration was measured and adjusted using Western blot lysis buffer.
SDS-PAGE and immunoblotting
Western blotting was performed as previously described.54 In brief, whole cell lysates were mixed with 4x or 6x Protein Sample Loading Buffer (LI-COR, at a final dilution of 1x) supplemented with 10% β-mercaptoethanol (Sigma Aldrich), heated at 95°C for 5 min, separated on NuPAGE 4±12% Bis-Tris Gels (Invitrogen) for 90minat 100 V and blotted onto Immobilon-FL PVDF membranes (Merck Millipore). The transfer was performed a constant voltage of 30 V for 30 minutes. After transfer, the membrane was blocked in 1% Casein in PBS (Thermo Scientific). Proteins were stained using primary antibodies against 229E N (1:2000), NL63 N (1:1000), OC43 N (1:1000), IFITM1 (1:1000,), IFITM2 (1:1000), IFITM3 (1:1000) ACE2 (1:1000), GAPDH (1:1000) and Infrared Dye labelled secondary antibodies (LI-COR IRDye). Band intensities were quantified using Image Studio (LI-COR).
Analysis of cell-associated SARS-CoV-2 N antigen
A total of 300,000 Calu3 cells per well were seeded in 12-well plate. At 18- and 64-hours post-seeding, cells were transfected with siRNAs targeting IFITM1, IFITM2 or IFITM3 or a non-targeting control siRNA using RNAimax according to the manufacturer’s instructions. At 24 hours after the first transfection and 6 hours after the second transfection, cells were treated with IFN-β at 500U/ml or left untreated. After the second round of IFN stimulation, cells were incubated on ice or at 37°C for 1h before infected with SARS-CoV-2 at MOI 2.5. Cells were further incubated on ice or at 37°C for 2h and then washed once by PBS and lysed for western blot analysis.
ACE2, CD46 and CD4 flow cytometry analysis
A549 stably expressing TMPRSS2 and ACE2, Hela stably expressing ACE2 or TZM-bl were transfected with expression constructs for IFITM1, 2 or 3 using TranslT-X2(Mirus Bio) and TranslT-LT1 (Mirus Bio) respectively according to manufacturer protocol. 24h later the medium was replaced and 48h posttransfection cells were harvested using Versene (Thermo Fischer) 15 minutes at 37°C. After washing with PBS, cells were stained with the eBioscience Fixable Viability Dye eFluor 780 (Thermo Fisher) for 15 minutes at room temperature in the dark. Afterwards cells were washed three times with PBS and, only where indicated, cells were permeabilized with 200μl of BD Cytofix/Cytoperm™ Fixation/Permeabilization Solution Kit (BD Biosciences) for 20 minutes at room temperature. Afterwards, cells were washed twice with 200μl of PBS or Perm/wash Buffer (BD Biosciences) and incubated with primary antibody (α-ACE2, α-CD46 conjugated with Alexa Fluor 647 or α-CD4 conjugated with PerCP-Cy5.5) diluted 1:200 in PBS or in Perm/Wash Buffer for 1hat 4°C. For the ACE2 staining, cells were washed three times with PBS or Perm/wash Buffer and stained with secondary antibody (Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™647) diluted 1:200 for 1hat 4°C. After washing three times with PBS or Perm/Wash Buffer, cells were resuspended in 100μl of PBS and analysed on BD FACS Canto II using BD FACS Diva and FlowJo software.
ACE2 flow cytometry analysis in Calu-3 cells
Calu-3 cells were seeded in a 6 well format. The next day, cells were transfected with either non-targeting or IFITM1, 2 or 3 siRNA following the siRNA protocol described above. In the indicated conditions, cells were simulated with 1000 U/ml IFN-β. 6h after the second transfection, cells were harvested using Versene (Thermo Fischer) 15 minutes at 37°C. Staining was performed as previously described.55 In brief, after washing with PBS cells were stained with the eBioscience Fixable Viability Dye eFluor 780 (Thermo Fisher) for 15 minutes at room temperature in the dark. Afterwards cells were permeabilized using 200μl of ice-cold methanol for 10 minutes, washed once with FACS buffer (2% FCS in PBS) and blocked with 200μl of FACS buffer on ice for 10 minutes. Then, cells were stained with primary antibody against ACE2 diluted 1:200 in FACS buffer and incubated at 4°C for 45 minutes. Cells were washed twice with FACS buffer and stained with secondary antibody (Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™647) diluted 1:200 in FACS buffer at 4°C. After 30 minutes, cells were washed twice with FACS buffer and resuspended in 100μl of PBS and analysed on BD FACS Canto II using BD FACS Diva and FlowJo software.
Proximity ligation assay
The proximity ligation assay (PLA) was performed as previously described.56 In brief, HeLa cells were seeded in a 24-well plate on a coverslip glass. 24 h postseeding the cells were transfected once withACE2 and either IFITM1, IFITM2 or IFITM3 flag tagged expression constructs. 24h post-transfection the media was replaced. 48h posttransfection, cells were washed twice with PBS and fixed 20 minutes with 4% PFA. Images were acquired on a Zeiss LSM 710 and processed using ImageJ (Fiji).
Immunofluorescence of A549 TMPRSS2 ACE2, Hela and Calu-3 cells
Cells were seeded on cover slides and the next day transfected with expression constructs for IFITM1, 2 or 3 using TranslT-LT1 (Mirus Bio) or TranslT-X2(Mirus Bio) according to manufacturer protocol. 48h later cells were fixed with 4% PFA for 20 minutes at room temperature, washed with PBS, permeabilized and blocked for 30 minutes at room temperature with perm/block solution containing 0.5% Triton X-100 and 5% FCS in PBS. Afterwards cells were stained 2h with primary antibody (α-ACE2 1:100, α-flag 1:100 and α-CD46 1:100) diluted in PBS in a wet chamber at room temperature. After washing with PBS-Tween 20, slides were incubated with secondary antibodies (Alexa Fluor IgG H+L 1:500) and 500 ng/ml DAPI 2h at 4°C. After washing with PBS-T and water, slides were mounted with Moviol-G. Images were acquired using a LSM 710 system.
Quantification and statistical analysis
Statistical analyses were performed using GraphPad PRISM 8 (GraphPad Software). P-values were determined using a two-tailed Student’s t test with Welch’s correction. Unless otherwise stated, data are shown as the mean of at least three independent experiments ±SEM. Significant differences are indicated as: ∗, p < 0.05; ∗∗, p < 0.01; ∗∗∗, p < 0.001; ∗∗∗∗, p < 0.0001. Statistical parameters are specified in the figure legends.
Acknowledgments
We thank Kerstin Regensburger, Jana-Romana Fischer, Birgit Ott, Martha Meyer, Regina Burger, Nicola Schrott and Daniela Krnavek for excellent technical assistance. The ACE2 vector and the SARS-CoV-2 S-HA plasmid were kindly provided by Shinji Makino and Stefan Pöhlmann. We thank Karl-Klaus Conzelmann and Stefan Pöhlmann for providing VSVΔG and Volker Thiel for providing the luciferase reporter CoV-229E construct. This study was supported by DFG grants to F.K. and K.M.J.S. (CRC 1279, SPP 1923), Baustein intramural funding to D.K., as well as the BMBF to F.K. and K.M.J.S. (Restrict SARS-CoV-2 and IMMUNOMOD). C.P.B., and S.N. are part of the International Graduate School in Molecular Medicine Ulm (IGradU).
Author contributions
Q.X. and C.P.B. performed most experiments with support by S.N. and D.K. L.E. and R.N. performed experiments with infectious SARS-CoV-1, SARS-CoV-2 and MERS-CoV assisted by F.Z. and D.N. M.V. performed PLA assays; J.H.L. performed co-immunoprecipitation assays. K.M.J.S. and C.D. provided resources and supervised some experiments. F.K. conceived the study, planned experiments, and wrote the manuscript. All authors reviewed, edited and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 13, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.106395.
Supplemental information
Data and code availability
-
•
Data: All data reported in this paper will be shared by the lead contact upon request
-
•
Code: This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Harris R.S., Hultquist J.F., Evans D.T. The restriction factors of human immunodeficiency virus. J. Biol. Chem. 2012;287:40875–40883. doi: 10.1074/jbc.R112.416925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kluge S.F., Sauter D., Kirchhoff F. SnapShot: antiviral restriction factors. Cell. 2015;163:774–774.e1. doi: 10.1016/j.cell.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 3.Malim M.H., Bieniasz P.D. HIV restriction factors and mechanisms of evasion. Cold Spring Harb. Perspect. Med. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rochman N.D., Wolf Y.I., Koonin E.V. Molecular adaptations during viral epidemics. EMBO Rep. 2022;23 doi: 10.15252/embr.202255393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauter D., Kirchhoff F. Key viral adaptations preceding the AIDS pandemic. Cell Host Microbe. 2019;25:27–38. doi: 10.1016/j.chom.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 6.Prelli Bozzo C., Kmiec D., Kirchhoff F. When good turns bad: how viruses exploit innate immunity factors. Curr. Opin. Virol. 2022;52:60–67. doi: 10.1016/j.coviro.2021.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Bailey C.C., Zhong G., Huang I.-C., Farzan M. IFITM-family proteins: the cell’s first line of antiviral defense. Annu. Rev. Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diamond M.S., Farzan M. Nature Publishing Group; 2013. The Broad-Spectrum Antiviral Functions of IFIT and IFITM Proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perreira J.M., Chin C.R., Feeley E.M., Brass A.L. IFITMs Restrict the Replication of Multiple Pathogenic Viruses. Academic Press. J Mol Biol. 2013;425:4937–4955. doi: 10.1016/j.jmb.2013.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi G., Schwartz O., Compton A.A. BioMed Central Ltd; 2017. More than Meets the I: The Diverse Antiviral and Cellular Functions of Interferon-Induced Transmembrane Proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith S.E., Weston S., Kellam P., Marsh M. Elsevier B.V.; 2014. IFITM Proteins - Cellular Inhibitors of Viral Entry. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li K., Markosyan R.M., Zheng Y.M., Golfetto O., Bungart B., Li M., Ding S., He Y., Liang C., Lee J.C., et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013;9:e1003124. doi: 10.1371/journal.ppat.1003124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai T.M., Marin M., Chin C.R., Savidis G., Brass A.L., Melikyan G.B. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amini-Bavil-Olyaee S., Choi Y.J., Lee J.H., Shi M., Huang I.C., Farzan M., Jung J.U. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe. 2013;13:452–464. doi: 10.1016/j.chom.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivanova P.T., Myers D.S., Milne S.B., McClaren J.L., Thomas P.G., Brown H.A. Lipid composition of viral envelope of three strains of influenza virus - not all viruses are created equal. ACS Infect. Dis. 2015;1:399–452. doi: 10.1021/acsinfecdis.5b00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ketter E., Randall G. Virus impact on lipids and membranes. Annu. Rev. Virol. 2019;6:319–340. doi: 10.1146/annurev-virology-092818-015748. [DOI] [PubMed] [Google Scholar]
- 17.Guo X., Steinkühler J., Marin M., Li X., Lu W., Dimova R., Melikyan G.B. Interferon-induced transmembrane protein 3 blocks fusion of diverse enveloped viruses by altering mechanical properties of cell membranes. ACS Nano. 2021;15:8155–8170. doi: 10.1021/acsnano.0c10567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao X., Guo F., Liu F., Cuconati A., Chang J., Block T.M., Guo J.T. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl. Acad. Sci. USA. 2014;111:6756–6761. doi: 10.1073/pnas.1320856111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shi G., Kenney A.D., Kudryashova E., Zani A., Zhang L., Lai K.K., Hall-Stoodley L., Robinson R.T., Kudryashov D.S., Compton A.A., Yount J.S. Opposing activities of IFITM proteins in SARS-CoV-2 infection. EMBO J. 2021;40 doi: 10.15252/embj.2020106501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wrensch F., Winkler M., Pöhlmann S. IFITM proteins inhibit entry driven by the MERS-Coronavirus Spike protein: evidence for Cholesterol-Independent Mechanisms. Viruses. 2014;6:3683–3698. doi: 10.3390/v6093683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang I.C., Bailey C.C., Weyer J.L., Radoshitzky S.R., Becker M.M., Chiang J.J., Brass A.L., Ahmed A.A., Chi X., Dong L., et al. Distinct patterns of IFITM-mediated restriction of filoviruses, SARS coronavirus, and influenza A virus. PLoS Pathog. 2011;7:e1001258. doi: 10.1371/journal.ppat.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prelli Bozzo C., Nchioua R., Volcic M., Koepke L., Krüger J., Schütz D., Heller S., Stürzel C.M., Kmiec D., Conzelmann C., et al. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat. Commun. 2021;12:4584. doi: 10.1038/s41467-021-24817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nchioua R., Schundner A., Kmiec D., Prelli Bozzo C., Zech F., Koepke L., Graf A., Krebs S., Blum H., Frick M., et al. SARS-CoV-2 variants of concern hijack IFITM2 for efficient replication in human lung cells. J. Virol. 2022;96 doi: 10.1128/jvi.00594-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi S., Sainz B., Corcoran P., Uprichard S., Jeong H. Characterization of increased drug metabolism activity in dimethyl sulfoxide (DMSO)-treated Huh7 hepatoma cells. Xenobiotica. 2009;39:205–217. doi: 10.1080/00498250802613620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo L., Dial S., Shi L., Branham W., Liu J., Fang J.-L., Green B., Deng H., Kaput J., Ning B. Similarities and differences in the expression of drug-metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab. Dispos. 2011;39:528–538. doi: 10.1124/dmd.110.035873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhong J., Gastaminza P., Cheng G., Kapadia S., Kato T., Burton D.R., Wieland S.F., Uprichard S.L., Wakita T., Chisari F.V. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pyrc K., Sims A.C., Dijkman R., Jebbink M., Long C., Deming D., Donaldson E., Vabret A., Baric R., van der Hoek L., Pickles R. Culturing the unculturable: human coronavirus HKU1 infects, replicates, and produces progeny virions in human ciliated airway epithelial cell cultures. J. Virol. 2010;84:11255–11263. doi: 10.1128/JVI.00947-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroeder S., Mache C., Kleine-Weber H., Corman V.M., Muth D., Richter A., Fatykhova D., Memish Z.A., Stanifer M.L., Boulant S., et al. Functional comparison of MERS-coronavirus lineages reveals increased replicative fitness of the recombinant lineage 5. Nat. Commun. 2021;12:5324. doi: 10.1038/s41467-021-25519-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pfefferle S., Schöpf J., Kögl M., Friedel C.C., Müller M.A., Carbajo-Lozoya J., Stellberger T., von Dall’Armi E., Herzog P., Kallies S., et al. The SARS-coronavirus-host interactome: identification of cyclophilins as target for pan-coronavirus inhibitors. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peiris J.S.M., Yuen K.Y., Osterhaus A.D.M.E., Stöhr K. The severe acute respiratory syndrome. N. Engl. J. Med. 2003;349:2431–2441. doi: 10.1056/NEJMra032498. [DOI] [PubMed] [Google Scholar]
- 31.Zhao X., Li J., Winkler C.A., An P., Guo J.T. IFITM genes, variants, and their roles in the control and pathogenesis of viral infections. Front. Microbiol. 2018;9:3228. doi: 10.3389/fmicb.2018.03228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Narayana S.K., Helbig K.J., McCartney E.M., Eyre N.S., Bull R.A., Eltahla A., Lloyd A.R., Beard M.R. The interferon-induced transmembrane proteins, IFITM1, IFITM2, and IFITM3 inhibit hepatitis C virus entry. J. Biol. Chem. 2015;290:25946–25959. doi: 10.1074/jbc.M115.657346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kenney A.D., Zani A., Kawahara J., Eddy A.C., Wang X.L., Mahesh K.C., Lu M., Thomas J., Kohlmeier J.E., Suthar M.S., et al. Interferon-induced transmembrane protein 3 (IFITM3) limits lethality of SARS-CoV-2 in mice. EMBO Rep. 2023;7:e56660. doi: 10.15252/embr.202256660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ziegler C.G.K., Allon S.J., Nyquist S.K., Mbano I.M., Miao V.N., Tzouanas C.N., Cao Y., Yousif A.S., Bals J., Hauser B.M., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035.e19. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boson B., Legros V., Zhou B., Siret E., Mathieu C., Cosset F.-L., Lavillette D., Denolly S. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J. Biol. Chem. 2021;296 doi: 10.1074/jbc.RA120.016175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naskalska A., Dabrowska A., Szczepanski A., Milewska A., Jasik K.P., Pyrc K. Membrane protein of human coronavirus NL63 is responsible for interaction with the adhesion receptor. J. Virol. 2019;93:e00355-19–e419. doi: 10.1128/JVI.00355-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Long S., Li S., Zhang X., Peng Q., Chen S., Wang J. ACE2 affects the expression and function of IFITM3 during SARS-CoV-2 pseudovirus infection in Vero E6 cells. Viral Immunol. 2022;35:653–662. doi: 10.1089/vim.2022.0042. [DOI] [PubMed] [Google Scholar]
- 41.Basile A., Zannella C., Marco M.D., Franci G., Galdiero M., Sanna G., Manzin A., Chetta M., Turco M.C., Rosati A., et al. A specific anti-IFITM2 antibody bars the way to SARS-CoV-2 entry into host cells. bioRxiv. 2022 doi: 10.1101/2022.08.04.502768. Preprint at. [DOI] [Google Scholar]
- 42.Hickford D., Frankenberg S., Shaw G., Renfree M.B. Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genom. 2012;13:155. doi: 10.1186/1471-2164-13-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanco-Melo D., Nilsson-Payant B.E., Liu W.-C., Uhl S., Hoagland D., Møller R., Jordan T.X., Oishi K., Panis M., Sachs D., et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045.e9. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Smith N., Péré H., Charbit B., Bondet V., Chenevier-Gobeaux C., et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science (New York, N.Y.). 2020;369:718–724. doi: 10.1126/science.abc6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grodzki M., Bluhm A.P., Schaefer M., Tagmount A., Russo M., Sobh A., Rafiee R., Vulpe C.D., Karst S.M., Norris M.H. Genome-scale CRISPR screens identify host factors that promote human coronavirus infection. Genome Med. 2022;14:10. doi: 10.1186/s13073-022-01013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu W.-L., Grotefend C.R., Tsai M.-T., Wang Y.-L., Radic V., Eoh H., Huang I.-C. Δ20 IFITM2 differentially restricts X4 and R5 HIV-1. Proc. Natl. Acad. Sci. USA. 2017;114:7112–7117. doi: 10.1073/pnas.1619640114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 48.Muth D., Meyer B., Niemeyer D., Schroeder S., Osterrieder N., Müller M.A., Drosten C. Transgene expression in the genome of Middle East respiratory syndrome coronavirus based on a novel reverse genetics system utilizing Red-mediated recombination cloning. J. Gen. Virol. 2017;98:2461–2469. doi: 10.1099/jgv.0.000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lawrenz J., Xie Q., Zech F., Weil T., Seidel A., Krnavek D., van der Hoek L., Münch J., Müller J.A., Kirchhoff F. Severe acute respiratory syndrome coronavirus 2 vaccination boosts neutralizing activity against seasonal human coronaviruses. Clin. Infect. Dis. 2022;75:e653–e661. doi: 10.1093/cid/ciac057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lemoine F., Correia D., Lefort V., Doppelt-Azeroual O., Mareuil F., Cohen-Boulakia S., Gascuel O. NGPhylogeny.fr: new generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019;47:W260–W265. doi: 10.1093/nar/gkz303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Letunic I., Bork P. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nchioua R., Kmiec D., Müller J.A., Conzelmann C., Groß R., Swanson C.M., Neil S.J.D., Stenger S., Sauter D., Münch J., et al. SARS-CoV-2 Is Restricted by Zinc Finger Antiviral Protein despite Preadaptation to the Low-CpG Environment in Humans. mBio. 2020;11 doi: 10.1128/mBio.01930-20. e01930–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koepke L., Winter B., Grenzner A., Regensburger K., Engelhart S., van der Merwe J.A., Krebs S., Blum H., Kirchhoff F., Sparrer K.M.J. An improved method for high-throughput quantification of autophagy in mammalian cells. Sci. Rep. 2020;10:12241. doi: 10.1038/s41598-020-68607-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherman E.J., Emmer B.T. ACE2 protein expression within isogenic cell lines is heterogeneous and associated with distinct transcriptomes. Sci. Rep. 2021;11 doi: 10.1038/s41598-021-95308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Volcic M., Sparrer K.M.J., Koepke L., Hotter D., Sauter D., Stürzel C.M., Scherer M., Stamminger T., Hofmann T.G., Arhel N.J., et al. Vpu modulates DNA repair to suppress innate sensing and hyper-integration of HIV-1. Nat. Microbiol. 2020;5:1247–1261. doi: 10.1038/s41564-020-0753-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data: All data reported in this paper will be shared by the lead contact upon request
-
•
Code: This paper does not report original code
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.