Extended Data Fig. 6. Characterization of aNB→S1 projections co-releasing ACh/GABA and their role in neuropathic pain.
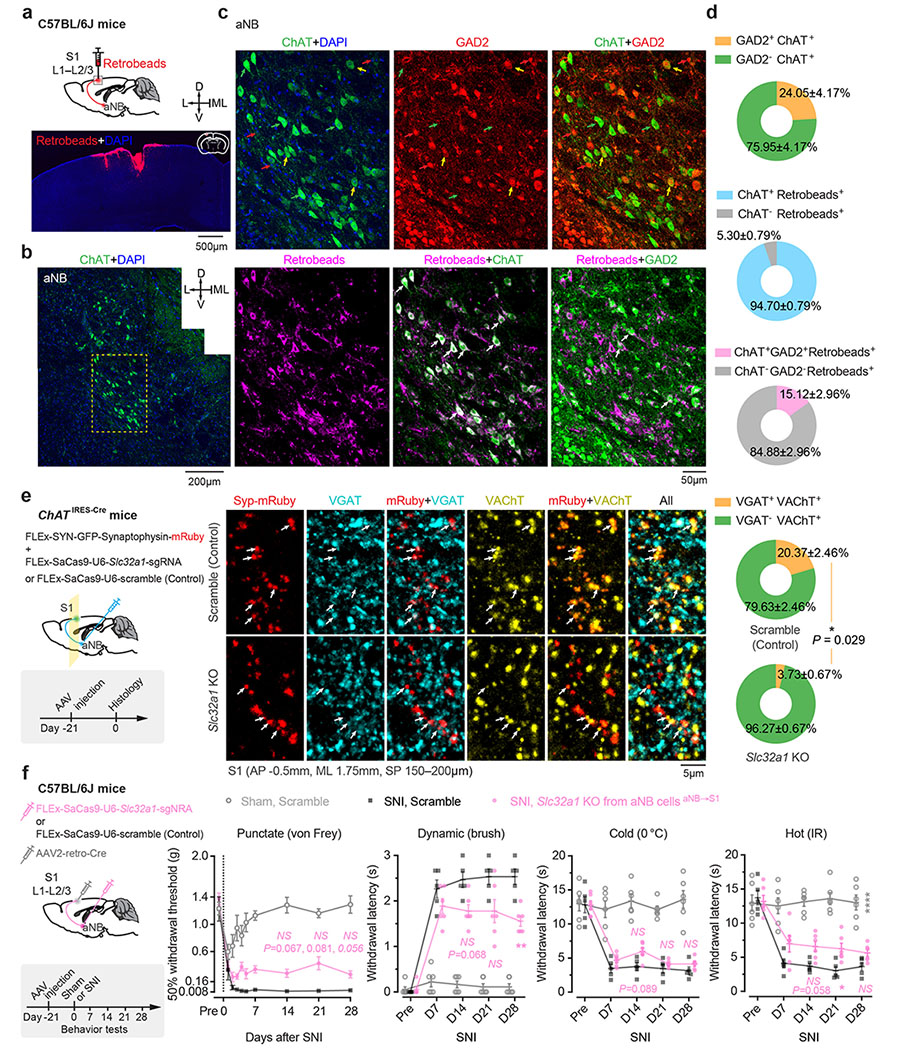
a, Schematic of experimental design. Retrobeads were injected into L1–L2/3 of S1 to trace S1-projecting cells in the aNB region (3 mice). b,c, Immunofluorescence images in the aNB region showing ChAT+ cells (b) and colocalization of ChAT, GAD2, and Retrobeads (c). d, Quantification of data shown in (c) (3 mice). Percentages of ChAT+ cells expressing GAD2, S1-projecting (Retrobeads+) cells expressing ChAT, or both ChAT and GAD2. e, Left, experimental design and timeline for CRISPR/Cas9-mediated deletion of Slc32a1 gene (encoding VGAT) in aNB cholinergic neurons and labeling of their axonal boutons with synaptophysin (Syp)-mRuby. Middle, immunofluorescence images showing colocalization of VGAT and VAChT in Syp-mRuby+ cholinergic boutons in S1. Right, percentages of VAChT+ boutons expressing VGAT in control and Slc32a1 knockout (KO) mice (n = 4 mice per group). Arrows, Syp-mRuby+ boutons. Scramble, a control vector without Slc32a1 sgRNA sequence. f, Left, experimental design for selective deletion of Slc32a1 gene in aNB–S1 projection neurons. Right, nociceptive thresholds under various conditions (n = 6, 5, 6 mice). VGAT KO in aNB–S1 projection neurons has no marked effects on SNI-induced mechanical and thermal allodynia in mice (punctate, P = 0.067, 0.081, 0.057; dynamic, 0.068, 0.19, 0.0093; cold, 0.089, 0.99, 0.82; hot, 0.058, 0.048, 0.16). Mean ± SEM. *P < 0.05, **P < 0.01; NS, not significant; by two-sided Mann-Whitney U test (e) or two-way ANOVA followed by Bonferroni’s test (f). See detailed statistics in Supplementary Table 1.
