Fig. 5.
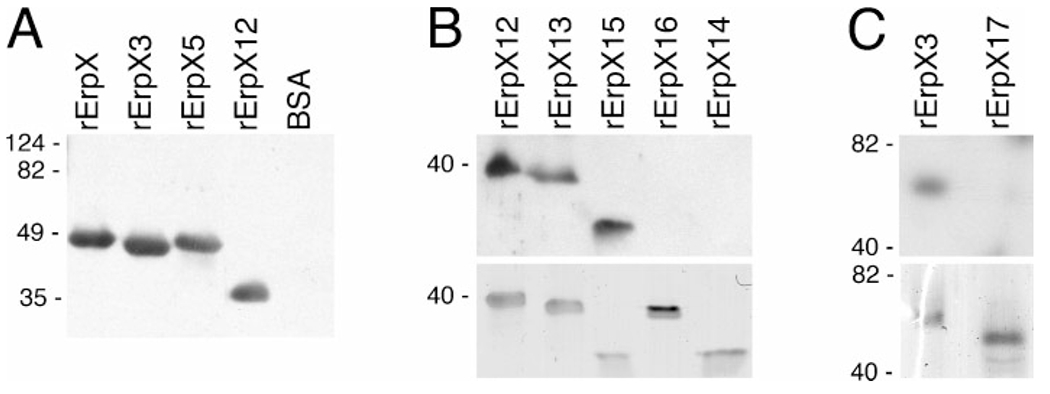
Ligand-affininty blot analysis of laminin binding by recombinant ErpX (rErpX) and truncated derivatives thereof. (A) Full-length ErpX and three of the examined truncations. The rightmost lane contained an equal molar content of BSA, to serve as a control for non-specific laminin binding. (B) The upper panel shows results of laminin-binding analysis of truncated proteins rErpX12, rErpX13, rErpX15, rErpX16 and rErpX14, and the lower panel shows the same amounts of each protein in a gel stained with Coomassie brilliant blue. Note that even though relatively less rErpX15 appears to have been loaded onto this gel, the laminin-binding signal for that protein was comparable to those from ErpX12 and rErpX13. (C) The upper panel illustrates results of laminin-binding analysis of proteins rErpX3 and rErpX17, and the lower panel shows the proteins stained with Coomassie brilliant blue. Migration positions of molecular mass standards are indicated on the left of each panel. Highly charged proteins such as ErpX often exhibit aberrant SDS-PAGE mobilities (Stevenson et al., 1998): note that rErpX-5 migrated at a rate suggestive of a molecular mass greater than rErpX-3, even though rErpX-5 is actually the smaller protein.
