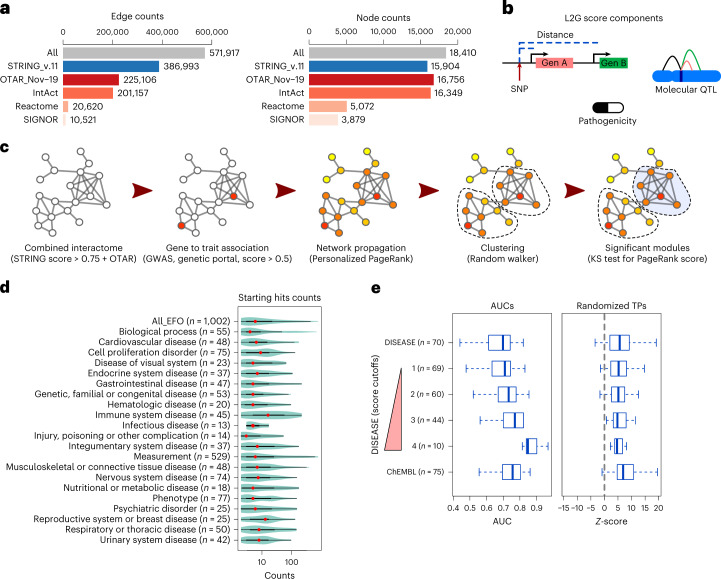
Fig. 1. Implementation and benchmarking of network-based augmentation of GWAS.
a, Edge and node counts of the combined interactome and its components. OTAR is the Open Targets combined physical protein interaction network that is provided via a Neo4j Graph Database. b, Graphic representation of some L2G components: SNP-to-gene distance, data from QTLs and variant effect predictions. The integration of information into the L2G score has been described previously11. c, Graphical representation of the network-based approach: network propagation of the initial input, clustering using a random walker to find gene communities and scoring of modules using the distribution of PageRank score. KS, Kolmogorov–Smirnov. d, Number of starting genes linked to traits, grouped in therapeutic areas. In the violin plot, the red dots represent the median, the limits of the thick line correspond to quartiles 1 and 3 (25% and 75% of the distribution) and the limits of the thin line are 1.5× the interquartile range. e, Benchmarking of the method, using as a starting signal genes from the Open Targets Genetics portal with a L2G score >0.5. AUC values are calculated using as positive hits the DISEASE database, with increasing cutoff values for its gene-to-trait score (Methods), as well as clinical trials data from the ChEMBL database (clinical phase II or higher). We also re-calculated the AUC values and determined Z-scores reflecting the deviation in AUCs relative to those observed after randomization of the list of true positives (TPs). In the boxplots, the middle lines represent the median, the limits of the box are quartiles 1 and 3 and the whiskers represent 1.5× the interquartile range.