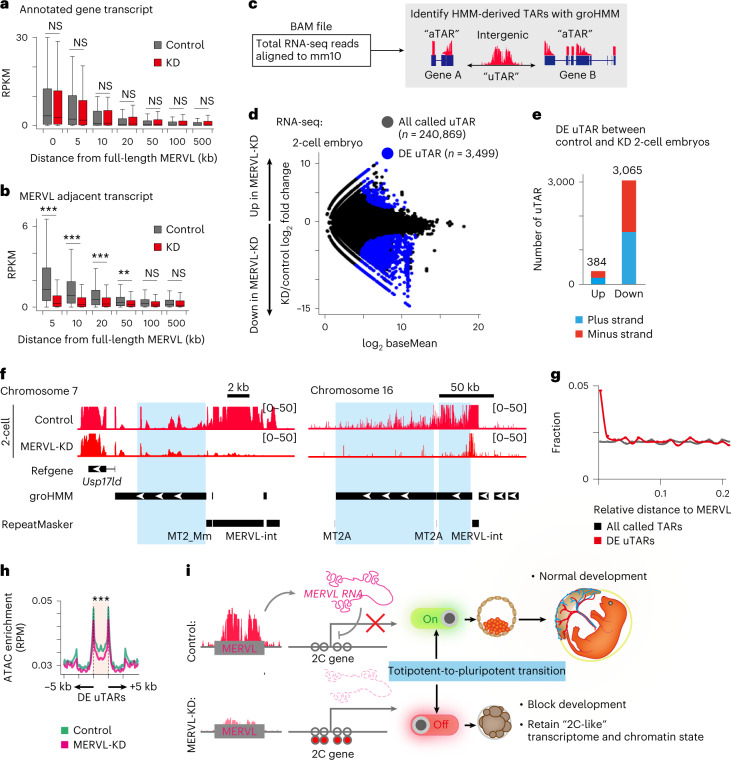
Fig. 6. Intergenic transcription from the adjacent regions to MERVL was interfered with by MERVL-KD.
a, Box plot showing the RPKM values for annotated genes, grouped by their distance to ASO-targeted full-length MERVL. Central bars represent medians, the boxes encompass 50% of the data points and the whiskers indicate 90% of the data points. NS, not significant, two-tailed unpaired t-tests. b, Box plot showing the RPKM values for adjacent transcripts grouped by their distance to ASO-targeted full-length MERVL. Central bars represent medians, the boxes encompass 50% of the data points and the whiskers indicate 90% of the data points. **P < 0.01, ***P < 0.001, two-tailed unpaired t-tests. c, Schematic of procedure for transcript calling with groHMM, a two-state HMM-based algorithm41. aTAR, annotated transcriptionally active regions (blue rectangles); uTAR, unannotated intergenic transcriptionally active region (two-headed black arrow). d, RNA-seq differential expression analysis of uTARs between control and MERVL-KD two-cell stage embryos. MA plot showing differentially expressed (DE) uTARs between control and MERVL-KD embryos. DE uTARs were defined with a Padj < 0.01 (binomial test with Benjamini–Hochberg correction) and shown in blue circles (n = 3,499). e, Number of upregulated (up)- and downregulated (down)-uTARs in MERVL-KD two-cell stage embryos from d. Each transcriptional direction in the DE uTARs is shown in blue (plus strand) and red (minus strand). f, Track views show RNA-seq signals in control and MERVL-KD two-cell stage embryos on two representative uTARs. DE uTARs are highlighted in blue. The y-axis represents normalized tag counts for total RNA-seq in each sample. g, Distributions of relative distances of DE uTARs (red) and all called TARs (black) to MERVL loci. h, Average tag density plots of ATAC-seq enrichment around DE uTARs (±5 kb) in control and MERVL-KD two-cell stage embryos. RPM, reads per million. ***Padj < 0.001, Mann–Whitney U-test with Bonferroni correction. i, Model: MERVL-mediated totipotent-to-pluripotent transition during preimplantation development. MERVL and adjacent intergenic regions are expressed in a zygote and two-cell-stage-specific manner and suppresses a subset of 2C gene expressions through an unknown mechanism to facilitate the totipotent-to-pluripotent transition. KD of MERVL transcript compromises these processes and results in retention of two-cell-like states and embryonic lethality. Data for panels a, b, d, e and g are available as source data.