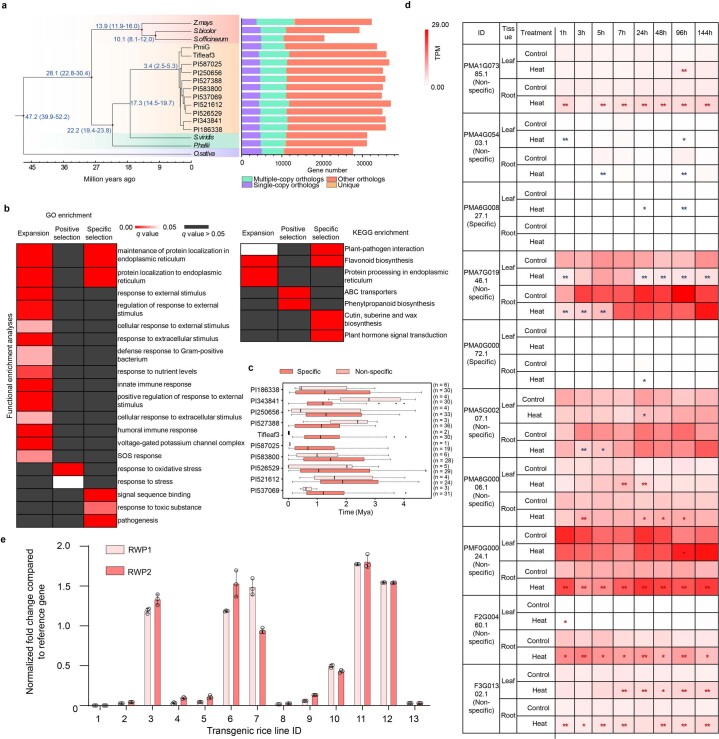
Extended Data Fig. 7. Expansion and expression of RWP-RK transcription factor (TF) family members.
a, Estimation of the divergence times of seven pearl millet accessions and six closely related species. PmiG represents Tift 23D2B1-P1-P5. The right panel displays the distribution of single-copy, multiple-copy, unique and other gene orthologs. b, Functional enrichment analyses showed the enrichment of some genes in stress-related processes or pathways among expanded family, specific, and positively selected gene sets. c, Estimated insertion times of LTR TEs encompassing the specific RWP-RKs of pearl millet belonging to clade A and the nonspecific RWP-RKs belonging to clade B. Center line, median; box limits, upper and lower quartiles; whiskers, 1.5x interquartile range; and dots represent outliers. d, Expression of seven RWP-RK genes under control (CK) and heat treatments (H). Significant differences were performed by DESeq2 to determine fold change (FC) of gene expressions between the CK and H groups (two-tailed Wald test). The FC was determined by * (0 < |log2FC|< 1, FDR-adjusted P < 0.05) or ** (|log2FC|>= 1, FDR-adjusted P < 0.05). The blue and red */** represent gene down- and upregulation, respectively. e, Expression of two RWP fragments (RWP1 and RWP2) in 13 transgenic rice lines. A value above 0 indicates the upregulation of this gene. Error bars, mean ± s.d.; n = 3 biological replicates.