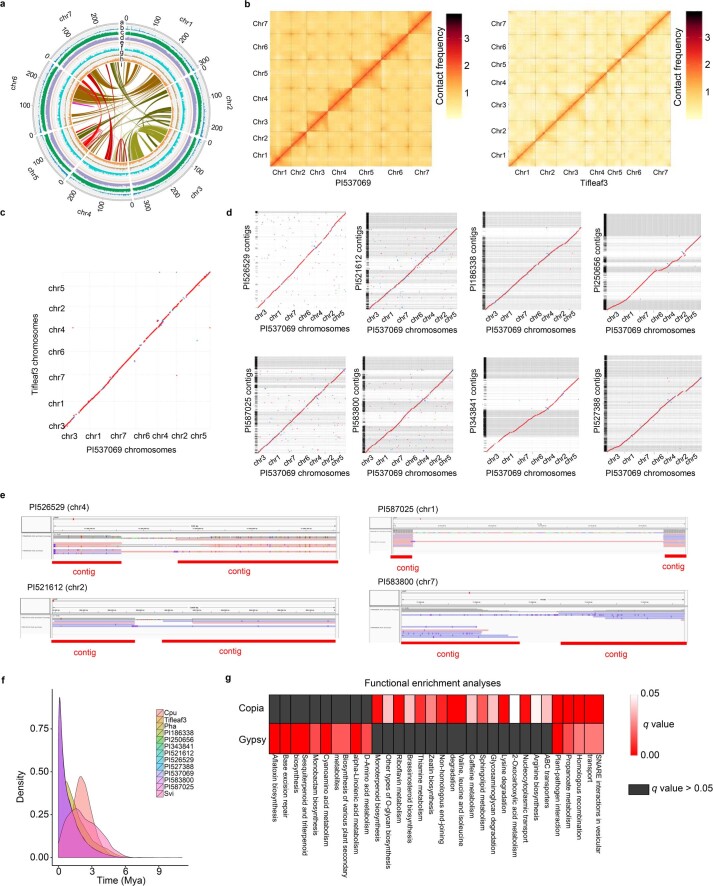
Extended Data Fig. 1. High quality assembled genomes in pearl millet.
a, Genome landscape of pearl millet (PI537069). Track ‘a’: the seven chromosomes at the Mb scale; track ‘b’: chromosomal distribution of gene models in which the gene density ranged from 201 to 503,365 bp/Mb; track ‘c’: chromosomal distribution of TEs for which the density was 58,334 to 945,223 bp/Mb; track ‘d’: TE/LTR distribution, ranging from 122,500 to 920,183 bp/Mb; track ‘e’: GC content along the assembled genome, which ranged between 20% and 60%/Mb; track ‘f’ and ‘g’: numbers of SNPs (5 to 27,243/Mb) and indels (1 to 5,952/Mb); track ‘h’: number of SVs (6 to 422/Mb); track ‘i’: collinear blocks of the pearl millet (PI537069) genome. b, Hi-C contact matrices of PI537069 and Tifleaf3 assemblies. c, Collinearity between the PI537069 and Tifleaf3 genomes. The contigs of Tifleaf3 were connected with Hi-C data only. This plot showed good synteny of the pseudochromosomes of Tifleaf3 and another assembly based on Hi-C data from PI537069. d, Synteny of eight contig-level assemblies with the PI537069 chromosome-level assembly. e, Assessment of the contig connections based on HiFi reads. Here the four examples show a long HiFi read across two contigs. f, Insertion times of LTRs in pearl millet and three related species, Cenchrus purpureus (Cpu), Panicum hallii (Pha) and Setaria viridis (Svi). g, Functional enrichment analyses of genes encompassed by intact Copia and Gypsy TEs.