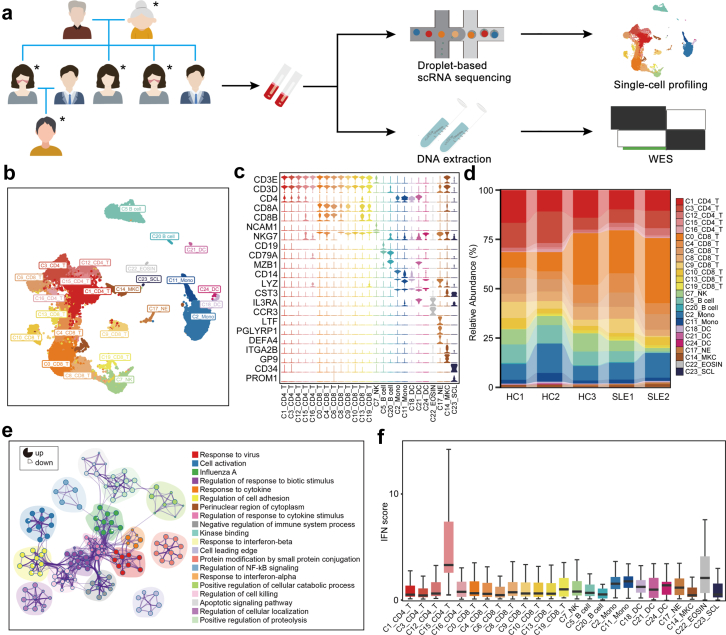
Fig. 1.
PBMCs profiling of a SLE family pedigree at the single-cell resolution. (a) Schematic map of experimental strategy. Individuals with butterfly erythema on their faces represent SLE patients, others represent HCs. The samples marked with ∗ are used to perform scRNA-seq and WES. (b) UMAP and clustering of 47,349 cells from 5 samples. (c) Violin plots depicting clusters are defined by a set of known marker genes. Heights denote average expression levels; widths denote cell densities. (d) Sankey plot representing cell abundance of each cluster (n = 25) across the 5 individuals (3 HCs and 2 SLE samples). (e) Network of enriched terms by the differentially expressed genes in T cells: colored by cluster ID. The proportion of the color in the circle represents the proportion of up-regulated genes in the term, and the proportion of the blank represents the proportion of down-regulated genes. (f) IFN scores across the clusters.