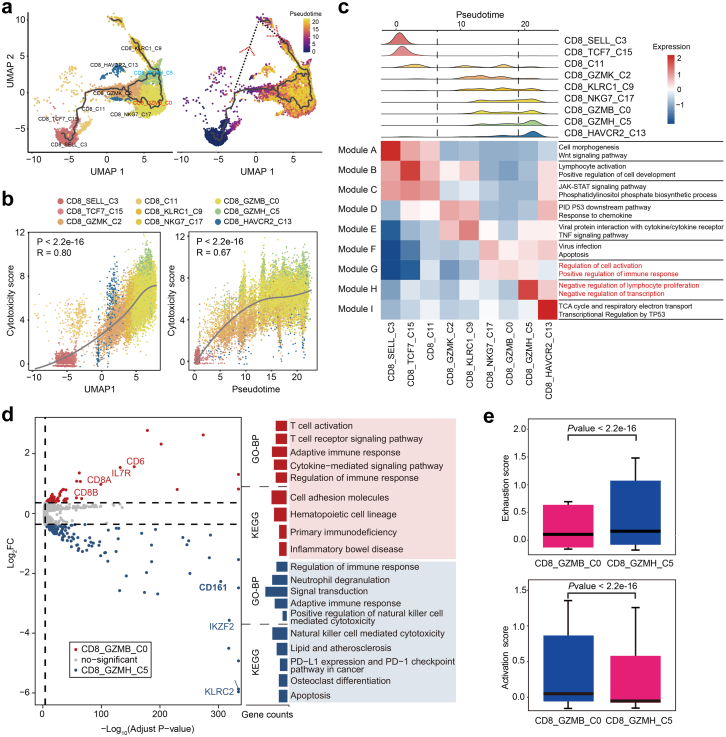
Fig. 3.
Analysis of CD8+T cell subsets transition trajectory in SLE. (a) Pseudotemporal analysis of CD8+ T cell subsets. Trajectory of CD8+ T Cell subclusters is inferred using monocle3 and subclusters are marked by colors (left). Pseudotime-ordered variables are inferred using monocle2 (right). Dotted lines and arrows indicate inferred differentiation trajectory and direction. (b) Correlation between pseudo-time and cytotoxicity of CD8+ T cell subsets. The solid line represents loess fitting of the relationship between cytotoxicity scores and Monocle components. P values are calculated by Spearman correlation. (c) Distribution of CD8+ subpopulations during the transition, along with the pseudo-time (upper). Heatmap showing the co-expression modules with the highest average expression in each CD8+ T cell subcluster (lower). (d) Volcano plots (left) and GO/KEGG enrichment analysis (right) of differentially expressed genes between CD8_GZMB and CD8_GZMH (left). Red represents upregulation in CD8_GZMB, and blue represents upregulation in CD8_GZMH. (e) Boxplot indicating the average expression of exhaustion and activation gene signatures in CD8_GZMB and CD8_GZMH. The P values are from a Wilcoxon test. Loess, locally weighted regression.