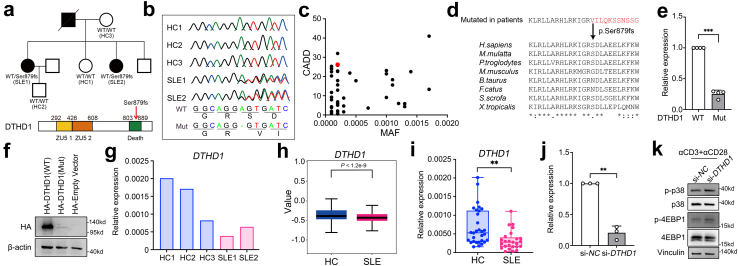
Fig. 5.
Loss of function of DTHD1 from SLE patients. (a) SLE family pedigree and schematic map showing mutation (Ser879fs) location within DTHD1 protein. (b) Sanger sequencing reads indicating variant location. (c) CADD scores versus MAF for the new Ser879fs patient-derived DTHD1 variant as compared with DTHD1 variants with an MAF cutoff of >10−4 from the gnomAD database. (d) Amino acid sequence alignment of DTHD1 death domain across species. The arrow indicates the position of the Ser589fs mutation (e and f) qPCR (e) and western blotting (f) analysis of DTHD1 expression in HEK293T cells transfected with HA-tagged WT DTHD1 or mutant DTHD1 expressing plasmid. P value is determined by Paired t-test, P = 0.000158. (g) qPCR analysis of DTHD1 expression in this SLE family pedigree. (h) Expression of DTHD1 in scRNA dataset between SLE samples and HCs. (i) qPCR analysis of DTHD1 expression in another cohort (HCs, n = 30; SLEs, n = 25). P value is determined by Unpaired t-test, P = 0.00178 (j) qPCR analysis of DTHD1 expression in primary CD8+ T cells electroporated with si-NC or si-DTHD1 siRNAs (50 nM). P value is determined by Paired t-test, P = 0.00608 (k) Western blotting analysis of p-p38, p38, p-4EBP1, 4EBP1 in primary CD8+ T cells electroporated with si-NC or si-DTHD1 siRNAs after treatment with αCD3 and αCD28. One representative experiment of three is shown (f and k). ∗∗P < 0.01, ∗∗∗P < 0.001.