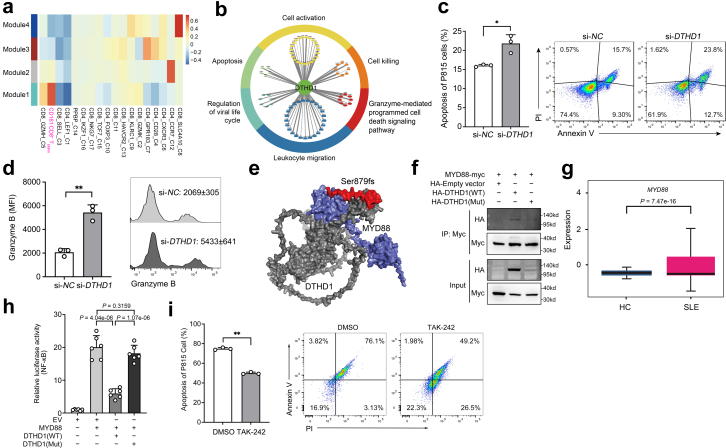
Fig. 6.
DTHD1 deficiency increased the cytotoxicity of CD161−CD8+TEMRAcells. (a) Heatmap showing the correlation of co-expression modules with different cell subsets. (b) Functional annotations of DTHD1-related genes. (c) Quantification (left) and flow cytometry (right) of the apoptosis percentage of P815 cells after cocultured with primary CD8+ T cells electroporated with si-NC or si-DTHD1 siRNAs for 5 h at the ratio of 1:5. P value is determined by Unpaired t-test, P = 0.0124. (d) Flow cytometric analysis of mean fluorescence intensity (MFI) of Granzyme B of CD161−CD8+ T cells electroporated with si-NC or si-DTHD1 siRNAs after coculture with P815 cell. P value is calculated by Unpaired t-test, P = 0.0012. (e) Structural prediction of DTHD1-MYD88 interaction by ZDOCK. Grey, DTHD1; blue, MYD88; red, mutated regions in DTHD1. (f) Western blotting analysis of HA in HEK293T cells co-transfected with myc-MYD88 and HA-empty vector or HA-DTHD1 (WT) or HA-DTHD1 (Mutant) expressing plasmids after immunoprecipitated with anti-Myc beads. (g) Boxplot indicating the average expression of MYD88 in CD161−CD8+ TEMRA cells in HCs and SLE samples from our dataset. P value is from Wilcoxon rank-sum test. (h) Luciferase activity analysis of lysates of HEK293T cells co-transfected luciferase reporter plasmid for NF-κB, pRL-TK-renilla-luciferase plasmid, MYD88 plasmid and WT-DTHD1 plasmid or mutant DTHD1 plasmid (n = 6). P values are determined by Unpaired t-test. (i) Quantification (left) and flow cytometry (right) of the apoptosis percentage of P815 cells after cocultured with primary CD8+ T cells pre-treated with DMSO or TAK-242 for 5 h at the ratio of 1:5. P value is determined by Unpaired t-test, P = 0.0024. One representative experiment of three is shown (f). ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.