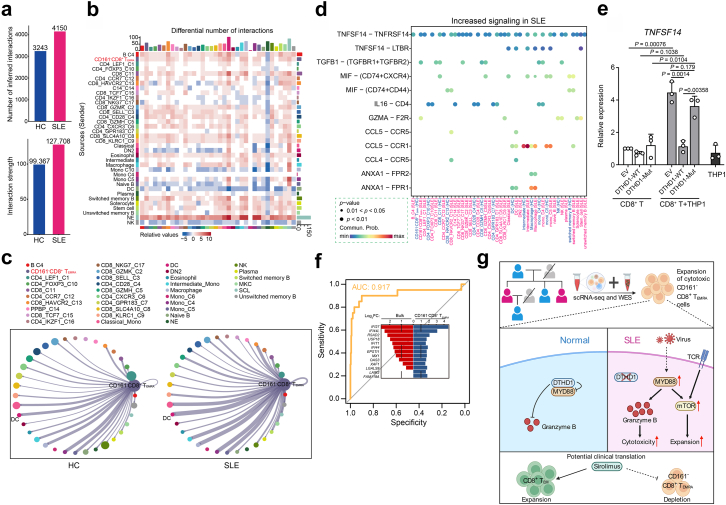
Fig. 7.
CD161−CD8+TEMRAcells contribute to SLE by LIGHT signaling. (a) The number (top) and strength (bottom) of interaction among all cells in HCs and SLE samples. (b) Heatmap of differential interactions between HCs and SLE samples in cell–cell communication network. The top bar indicates the sum of incoming signaling and right bar indicates the sum of outgoing signaling. Red indicates increased signaling and blue indicates decreased signaling in SLE. (c) Number of significant ligand-receptor pairs between CD161−CD8+ TEMRA cells (outgoing) and other cell subclusters (incoming) in HCs (left) and SLEs (right). The relative number of ligand-receptor pairs is represented by the edge width. (d) Comparison of the significant outgoing signaling from CD161−CD8+ TEMRA cells between HC and SLE. Empty space means the communication probability is zero. P-values are computed from one-sided permutation test. ∗∗∗P < 0.001. (e) qPCR analysis of TNFSF14 expression in primary CD8+ T cells electroporated with empty vector, DTHD1-WT or DTHD-mut plasmid cocultured with THP1 cells (n = 3). P values are determined by unpaired t-test. (f) Receiver operating curve for out-of-sample prediction of case–control state by a logistic regression model trained on DEGs in CD161−CD8+ TEMRA cells. DEGs include IFI27, IFI44L, RSAD2, IFI44, FAM118A, LGALS9, MX1, EPSTI1, USP18, OAS3, LAIR2, IFIT1, XAF1. Inset depicts the changes of DEGs in the public transcriptome profile and CD161−CD8+ TEMRA cells. (g) Graphic abstract showing the expansion of CD161−CD8+ TEMRA in patients with SLE and DTHD1 downregulation promotes MYD88-mediated expansion and cytotoxicity of this pathogenic CD161−CD8+ TEMRA subset in SLE.