Abstract
Purpose:
Alpelisib is a PI3K alpha (PI3Kα)-selective inhibitor approved for the treatment of hormone receptor–positive/HER2-negative (HR+/HER2−) PIK3CA-mutated advanced breast cancer (ABC) based on the SOLAR-1 trial, which defined 11 substitutions in exons 7, 9, and 20 in PIK3CA (SOLAR1m). We report alpelisib effectiveness for ABC harboring SOLAR1m, as well as other pathogenic PIK3CA mutations (OTHERm) using comprehensive genomic profiling (CGP).
Experimental Design:
A total of 33,977 tissue and 1,587 liquid biopsies were analyzed using hybrid capture–based CGP covering the entire coding sequence of PIK3CA. Clinical characteristics and treatment history were available for 10,750 patients with ABC in the deidentified Flatiron Health-Foundation Medicine clinico-genomic database (FH-FMI CGDB).
Results:
PIK3CAm were detected in 11,767/33,977 (35%) of tissue biopsies, including 2,300 (7%) samples with OTHERm and no SOLAR1m. Liquid biopsy had 77% sensitivity detecting PIK3CAm, increasing to 95% with circulating tumor DNA fraction ≥2%. In patients with HR+/HER2− ABC and PIK3CAm receiving alpelisib/fulvestrant (ALP+FUL; n = 182) or fulvestrant alone (FUL; n = 119), median real-world progression-free survival (rwPFS) was 5.9 months on ALP+FUL [95% confidence interval (CI): 5.1–7.4] versus 3.1 months on FUL (95% CI: 2.7–3.7; P < 0.0001). In patients with OTHERm, median rwPFS was 4.0 months on ALP+FUL (95% CI: 2.8–10.1) versus 2.5 months on FUL (95% CI: 2.2–3.7; P = 0.0054).
Conclusions:
CGP detects diverse PIK3CAm in a greater number of patients with ABC than PCR hotspot testing; 20% of patients with PIK3CAm do not have SOLAR1m. These patients may derive benefit from alpelisib.
Translational Relevance.
Alpelisib is an α-selective inhibitor of PI3K, approved in combination with endocrine therapy (ET) for the treatment of hormone receptor–positive/HER2-negative advanced breast cancer (ABC) harboring one of 11 substitutions in exons 7, 9, and 20 of the PIK3CA gene. This article validates the SOLAR-1 trial findings in a real-world cohort, showing significant benefit of alpelisib/fulvestrant compared with fulvestrant alone in patients with PIK3CA-mutated ABC. Importantly, we observed substantial benefit in the patients with PIK3CA mutations outside of the approved list of 11 substitutions, accounting for 20% of PIK3CA-mutated breast cancer. These findings support the use of alpelisib in combination with ET to treat patients with metastatic breast cancer harboring activating mutations across the PIK3CA gene, including the C-terminus (exon 20), the helical domain (exon 9), the p85 binding domain (exon 1), the C2 domain (exon 4), and exon 13.
Introduction
PI3Kα, which is encoded by the PIK3CA gene, is a subunit of PI3K, a membrane-associated kinase that functions in signaling pathways that control cell growth and survival. PIK3CA mutations are common in several malignancies, including breast cancer (1), and are associated with poor outcomes and chemoresistance in patients with advanced hormone receptor–positive/HER2-negative (HR+/HER2−) breast cancer (2). Alpha-isoform selective inhibitors, such as alpelisib (BYL719) and taselisib (GDC-0032; ref. 3), as well as pan-PI3K inhibitors buparlisib (BKM120), copanlisib (BAY80-6946), and pictlisib (GDC-0941) have been investigated in combination with endocrine therapy (ET) as treatment for advanced breast cancer (ABC) with activating PIK3CA mutations.
SOLAR-1 (NCT02437318) was a randomized phase III trial that demonstrated significant improvement in progression-free survival (PFS) from the combination of alpelisib plus fulvestrant (ALP+FUL) compared with fulvestrant (FUL) alone in patients with PIK3CA mutation (PIK3CAm) HR+/HER2− ABC who had been previously treated with ET (4). Patients with ≥1 of 11 prespecified PIK3CA substitution mutations detected in a tissue biopsy were eligible to enroll, and no predictive value of specific PIK3CA mutations was observed (5). The FDA-approved ALP+FUL for this population, along with companion diagnostic (CDx) tests for tissue and liquid biopsies: the therascreen PIK3CA RGQ PCR kit (specifically designed to detect the 11 mutations) and next-generation sequencing (NGS) via FoundationOne CDx and FoundationOne Liquid CDx, which reports a wide spectrum of activating variants in the PIK3CA gene but includes only the 11 substitutions that CDx claims for alpelisib (6–8).
There are several questions relevant to clinical care that remain unaddressed in most guidelines for treating physicians:
(i) What relevance do mutations predicted to activate PIK3CA have when they fall outside of the defined set of 11 mutations of the companion diagnostic/SOLAR-1 trial definition? Is ALP use appropriate in patients with these mutations?
(ii) Is there a benefit to comprehensive genomic profiling (CGP) with NGS over PCR hotspot testing?
(iii) What is the sensitivity of liquid biopsies for detection of PIK3CA mutations, and what should be taken into consideration when evaluating a negative liquid biopsy?
Our study evaluates data from patients who underwent genomic profiling during routine clinical care to address these questions.
Materials and Methods
CGP
CGP results reported during routine clinical care were used in this study. Approval for this study, including a waiver of informed consent and Health Insurance Portability and Accountability Act waiver of authorization, was obtained from the Western Institutional Review Board (protocol 20152817). Studies were conducted in accordance with the Declaration of Helsinki.
Tissue biopsies from 33,977 patients with ABC were analyzed using NGS assays FoundationOne (F1) or FoundationOne CDx (F1CDx). The pathologic diagnosis of each case was confirmed on routine hematoxylin and eosin–stained slides, and all samples forwarded for DNA extraction contained a minimum of 20% tumor nuclei. Liquid biopsies from 1,587 patients were analyzed using FoundationOneLiquid CDx (F1LCDx). F1, F1CDx, and F1LCDx are validated hybrid capture–based CGP assays (9–11) performed in a Clinical Laboratory Improvement Amendment–certified, College of American Pathologists–accredited, New York State–approved laboratory (Foundation Medicine).
CGP was performed on hybridization-captured, adaptor ligation-based libraries for at least 324 cancer-related genes plus select introns from at least 28 genes frequently rearranged in cancer, including the full coding region of PIK3CA. Results were analyzed for base substitutions, short insertions/deletions (indels), rearrangements, and copy-number alterations.
For patients with multiple submitted biopsies of the same type, a single sample with the highest sequencing quality metrics was included. For tissue/liquid concordance analysis, a convenience cohort of 206 patients with F1CDx and F1LCDx results was analyzed. A CONSORT diagram for the Foundation Medicine genomics database analyses is provided in Supplementary Fig. S1.
Clinico-genomic data
This study used the nationwide (U.S.-based) deidentified Flatiron Health-Foundation Medicine clinico-genomic database (FH-FMI CGDB). The deidentified data originated from approximately 280 U.S. cancer clinics (∼800 sites of care). Retrospective longitudinal clinical data were derived from electronic health record (EHR) data, comprising patient-level structured and unstructured data, curated via technology-enabled abstraction, and were linked to genomic data derived from FMI CGP tests in the FH-FMI CGDB by deidentified, deterministic matching. Institutional Review Board approval of the study protocol was obtained prior to study conduct and included a waiver of informed consent (WCG IRB).
Real-world cohort generation
This study cohort consisted of all patients with confirmed diagnosis of ABC included in FH-FMI CGDB who had tissue or liquid biopsies profiled between January 2011 and December 2021 (10,750 patients at data cutoff of December 31, 2021). Patients with HR+/HER2− ABC and substitution or indels in PIK3CA predicted to be activating (SOLAR1m and/or OTHERm) receiving FUL alone or ALP+FUL in treatment line 2 or later were considered for the real-world validation of SOLAR-1 trial results (2,977 patients). To minimize bias introduced by missing treatment data, patients with a metastatic diagnosis date >90 days prior to their first visit in FH network, as well as patients with FMI report dates >60 days from their last recorded FH visit, were excluded. Only patients whose receptor status was known before treatment of interest started were evaluated. To ensure cohort independence, patients who received both FUL and ALP+FUL were excluded from the analysis. Finally, only the patients with disease progression data were included. Application of these criteria resulted in a cohort of 182 ALP+FUL and 119 FUL patients eligible for further treatment outcome analysis.
For the descriptive analysis of survival outcomes in patients without SOLAR1m, patients with ABC of any receptor subtype, bearing only non-SOLAR1 PIK3CA mutations (OTHERm) and treated with ALP in any combination were considered. To account for missing treatment data, patients with FMI report date >60 days from their last recorded FH visit were excluded. In this manner, a cohort of 77 eligible patients was selected.
A CONSORT diagram for the FH-FMI CGDB analyses is provided in Fig. 2. For further details about data analysis and endpoints, please refer to Supplementary Materials and Methods.
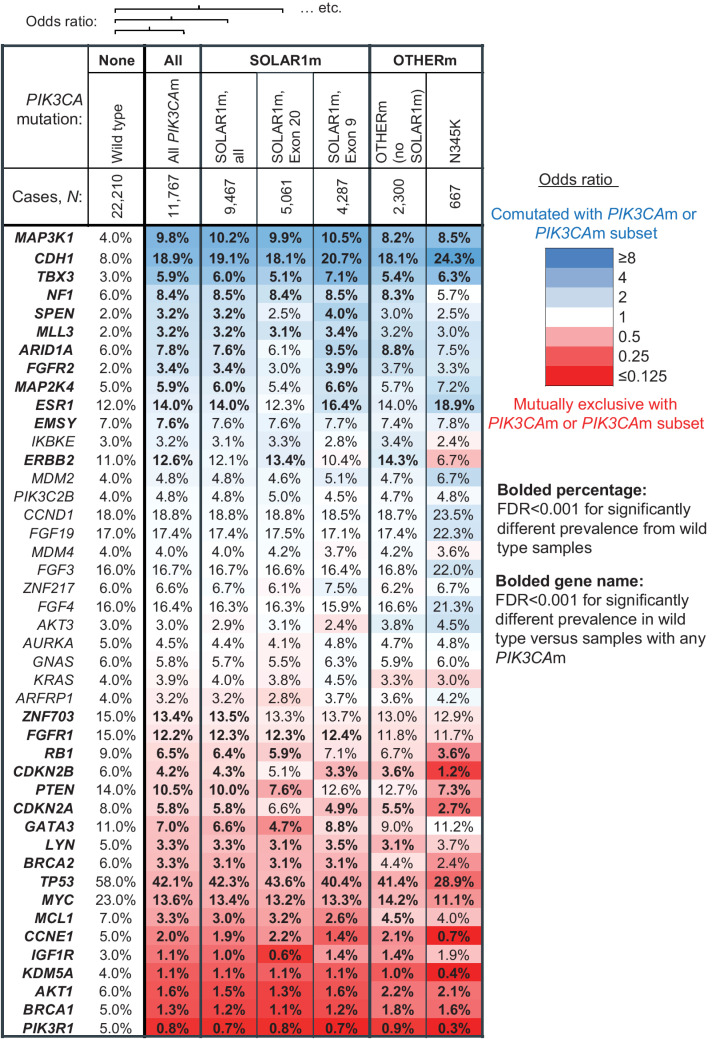
Figure 2.
The mutational landscapes of breast cancer tissue biopsies with SOLAR1m and OTHERm are similar. Co-occurrence and mutual exclusivity of other gene alterations with PIK3CA mutations in 33,977 tissue biopsies from patients with breast cancer. Mutations, rearrangements, and copy-number alterations were considered. Genes with ≥3% prevalence of genomic alteration in the wild-type and/or PIK3CA-mutated (PIK3CAm) cohort are arranged in order of OR. Heatmap colors correspond to degree of coalteration (blue) or mutual exclusivity (red) with PIK3CA mutations, as quantified by the log2 OR of being detected in the same sample as a PIK3CA mutation, or a particular type of PIK3CA mutation. The cohorts compared with PIK3CA wild-type samples are: all PIK3CAm, all SOLAR1m, the SOLAR1m exon 20 subset, the SOLAR1m exon 9 subset, the OTHERm cohort, and the N345K subset of the OTHERm cohort. Prevalence of mutations in each cohort indicated in each cell, and bolded where FDR < 0.001. P values were multiple test corrected across all genes assayed (including those not displayed) using the Benjamini–Hochberg method.
Survival analyses
Data analysis was conducted in RStudio (R version 4.1.0). The main endpoint in this study was real-world PFS (rwPFS). rwPFS was defined as a measure of time from the beginning of oncologist-defined, rule-based line of therapy (LOT) to either the date of real-world progression (rwP) or death. Because of the nature of real-world data (RWD), alternatives to RECIST are required to identify rwP. FH utilizes RWD collected in the EHR as part of routine clinical care to identify evidence of a patient's cancer worsening. Real-world overall survival (rwOS) was an additional endpoint of interest. In FH-FMI CGDB, rwOS is defined as the time from an index date to the date of death for individual patients who have died. Deaths are identified using FH mortality information, which has demonstrated high sensitivity, specificity, and accuracy as compared with the National Death Index. In this study, patients with missing record of progression or death were treated as right censored.
The two groups receiving FUL and ALP+FUL were compared on multiple demographic, clinical, and genomic characteristics. For numeric variable comparison (e.g., age at diagnosis), Kruskal–Wallis test was used. χ2 test was used for categorical variables (e.g., tumor type). To control for the rate of type I errors in null hypothesis testing when conducting multiple comparisons, the FDR method was applied.
Kaplan–Meier time-to-event analysis was used to obtain estimates of median rwPFS and survival curves with minimal bias. Random forest algorithm was used for multiple imputation of missing data. A logistic regression model including metastasis-free interval (MFI), tumor type, biopsy type, metastases sites, tumor mutational burden (TMB), PTEN mutation presence, prior chemotherapy, prior CDK 4/6 inhibitor therapy and LOT number was used to estimate individual propensity scores (Supplementary Fig. S7). Predicted probabilities of being assigned to ALP+FUL group were used for inverse probability of treatment weighting (IPTW) to achieve a balanced distribution of confounders across treatment groups and obtain less biased estimation of the treatment effect. To compare the survival distributions between the FUL and ALP+FUL groups, log-rank test was used.
For rwOS analysis, we used risk set adjustment, with the date of LOT start as index date and the date FMI report was received as entry date. Independent delayed entry assumption met: HR = 1.004 [95% confidence interval (CI), 0.99–1.017], P = 0.548, which indicated that hazard of death did not depend on when sequencing happened.
To describe individual subject's patterns of response to ALP treatment in the OTHERm group, we combined a swimmer plot with a table, containing relevant information about each patient's clinical and genomic background. The final graph included a bar showing the length of rwPFS in months for each patient, color coded by treatment type and progression indicators.
Circulating tumor DNA fraction estimation in liquid biopsy
The levels of circulating tumor DNA (ctDNA) shed for each liquid biopsy was quantified by calculating an investigational tumor fraction (TF), which merges two methods for estimation. When TF is elevated (≥10%), an estimate is returned on the basis of a measure of tumor aneuploidy that incorporates observed deviations in coverage across the genome. When lack of detectable tumor aneuploidy limits the ability to estimate TF, a variant-based calculation is made by identifying the highest allele fraction nongermline variant, excluding specific clonal hematopoiesis–associated alterations. This aneuploidy-based approach avoids erroneous inference of elevated TF due to the presence of germline variants detected at high variant allele frequency (VAF).
Data availability
The data supporting the findings of this study originated from Foundation Medicine, Inc and Flatiron Health, Inc. These deidentified data may be made available upon request, and are subject to a license agreement with FH and FMI; interested researchers should contact the corresponding author and cgdb-fmi@flatiron.com to determine licensing terms.
Results
Distribution of pathogenic mutations along the PIK3CA gene
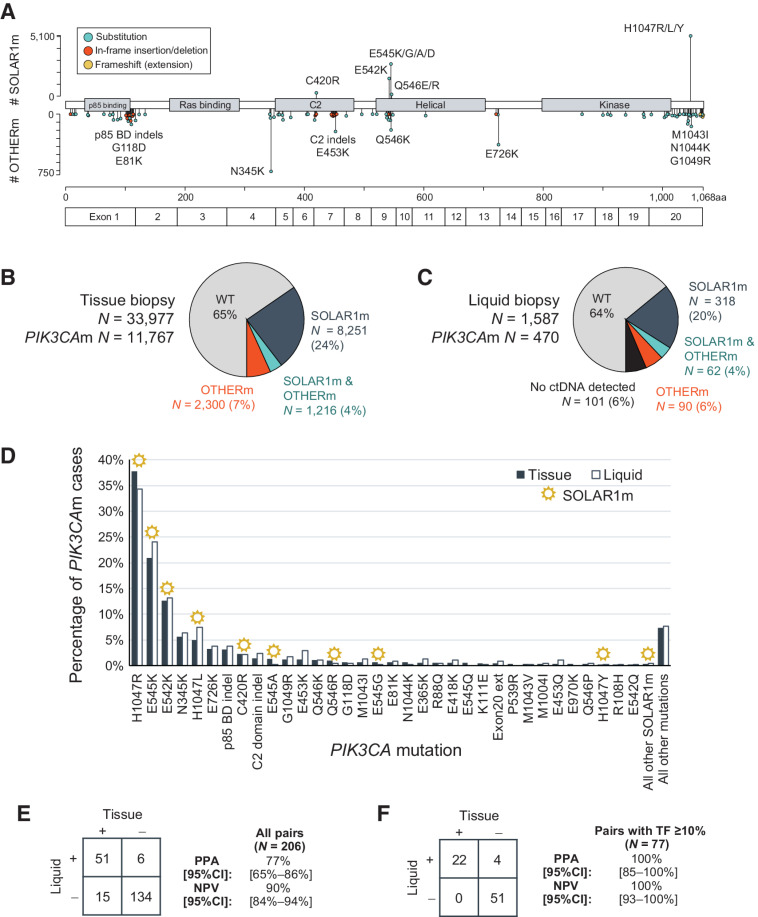
The 11 substitution mutations defined in the SOLAR-1 trial and which form the basis of PCR hotspot tests such as therascreen RGQ are found in the C2 domain (C420R), the helical domain (E542K, E545K/G/A/D, Q546E/R), and in the C-terminal tail (H1047R/L/Y; Fig. 1A). The full spectrum of pathogenic mutations identified by CGP among 33,977 tissue biopsies showed clusters in every domain but the Ras-binding domain and included not only a diverse collection of substitutions, but indels as well (Fig. 1A). In-frame indels clustered in the domain required for binding of p85 (the regulatory subunit of the kinase encoded by PIK3R1) and in the C2 domain. A set of frameshift mutations at very end of the protein function as extensions that activate the kinase by increasing its affinity to the plasma membrane (ref. 12; Fig. 1A). For simplicity, indels were grouped together according to domain in subsequent analyses.
Figure 1.
Prevalence of SOLAR1m and OTHERm in patients with breast cancer. A,PIK3CA mutations identified among 33,977 breast cancer tissue biopsies. Lollipops pointing upward correspond to the 11 substitution mutations defined in the SOLAR-1 trial (SOLAR1m). All other known and likely pathogenic mutations and indels are represented by lollipops pointing downward. Color coding denotes different variant types. An amino acid number ruler and exon demarcations are provided at the bottom. B, Pie chart summarizing the percentages of patients with PIK3CA mutations, as detected in tissue biopsies. “SOLAR1m & OTHERm” denotes at least 1 SOLAR1m and at least 1 OTHERm detected in the same biopsy. C, As in B, as detected in liquid biopsies, also denoting the percentage of patients without detectable ctDNA content. D, Detection of specific substitution mutations, deletions (p85 binding and C2 domains), and exon 20 extensions (frameshift or nonstop) by tissue and liquid biopsies. SOLAR1m are denoted with a sun icon. “All other mutations” denotes other pathogenic mutations that were detected at lower rates. E, Detection of PIK3CA mutations in tissue and liquid biopsies from 206 patients with both CGP results available. Sensitivity (PPA) and NPV were calculated with tissue as standard. F, As in E, but for the subset of 77 pairs where the liquid biopsy TF was ≥10%.
PIK3CA mutation detection in tissue and liquid biopsies
PIK3CA mutations were detected in 11,767/33,977 (35%) of tissue biopsies: 8,251 (24%) had only SOLAR1m detected, 2,300 (7%) had only OTHERm detected, and 1,216 (4%) harbored mutations of both types. In total, 19.5% (2,300/11,767) of tissue biopsies with PIK3CA mutations detected by CGP would have been missed by PCR testing designed to exclusively detect mutations defined in the SOLAR-1 trial (Fig. 1B).
ABC liquid biopsies contained detectable ctDNA in 94% of cases (1,486/1,587). Among liquid biopsies with ctDNA content, 42% (622/1,486) had PIK3CA mutations detected, with ratios of SOLAR1m and OTHERm comparable with tissue (32% SOLAR1m, 6% OTHERm, 4% both; Fig. 1C).
Several SOLAR1m made up the most prevalent PIK3CA mutations in both the tissue and liquid cohorts: H1047R (38% of tissue; 34% of liquid biopsies with PIK3CA mutations), E545K (21%; 24%), E542K (13%; 13%). However, some of the 11 substitutions were vanishingly rare: E545D was detected in 14 tissue samples and one liquid sample (0.12% of tissue; 0.21% of liquid biopsies with PIK3CA mutations), Q546E (0.20%; 0.21%), and H1047Y (0.27%; 0.21%). In contrast, several OTHERm were quite prevalent in PIK3CAm ABC: N345K (5.7% of tissue; 6.4% of liquid biopsies with PIK3CA mutations), E726K (3.2%; 3.8%), and indels in the p85 binding domain (3.1%; 3.8%; Fig. 1D).
Concordance of tissue and liquid biopsy detection of PIK3CA mutations
A cohort of 206 patients with both tissue and liquid biopsy CGP results ordered in the course of routine clinical care was used for concordance analysis between the two platforms (median collection time difference was liquid specimen collected 12 months after tissue specimen, interquartile range: 1.2–27 months). There was 77% positive percent agreement (PPA) at the patient level and 75% PPA at the variant level with tissue biopsy taken as truth (Fig. 1E). Liquid biopsy detected 43 SOLAR1m and 16 OTHERm concordantly with tissue, detected five SOLAR1m and six OTHERm that had not been detected in tissue, and missed detection of 12 SOLAR1m and five OTHERm (Supplementary Fig. S3). The 20 PIK3CA mutations detected in tissue but not in liquid tended to have lower VAFs in the tissue biopsy (median VAF 15%) compared with concordantly detected variants (median VAF 39%; Supplementary Fig. S3).
Among the paired samples, PIK3CA mutation detection by liquid increased to 95% PPA when the ctDNA fraction in the liquid biopsy was at least 2%—a threshold that still included 78% of the paired cohort (Supplementary Fig. S4). In the 77 samples with ctDNA fraction ≥10% (37% of the 206 paired cohort), the sensitivity of PIK3CA mutation in liquid detection was 100% (Fig. 1F). The negative predictive value (NPV) of a liquid biopsy was 90% in all 206 pairs, and 100% in the 77 pairs with ctDNA fraction ≥10% (Fig. 1E and F).
Comparison of PCR and CGP for PIK3CA mutation detection
We examined a subset of 128 patients who had both tissue CGP and tissue PCR hotspot testing results [therascreen RGQ PCR, which is designed to detect the 11 SOLAR1m (13)]. Thirty patients positively detected as having a PIK3CAm tumor by PCR were all also detected by CGP. Thirteen patients had an indeterminate PCR result, and 5 of these were detected as PIK3CAm by CGP. One patient who tested negative by PCR had a SOLAR1m identified by CGP. Among the 72 patients with a negative PCR result, CGP detected OTHERm in 7 patients (Supplementary Fig. S5A).
In summary, the total sensitivity of PCR with CGP as truth was 86% for SOLAR1m and 70% for all PIK3CAm (0% for OTHERm, by design). No patients detected by PCR were missed by CGP (Supplementary Fig. S5B).
OTHERm as oncogenic drivers or as secondary mutations
We examined the types of OTHERm to investigate whether certain mutations may be stronger oncogenic drivers than others. Mutations that tended to be subclonal in tumor biopsies included glutamate to lysine changes E970K, E365K, E453K, E81K, and E726K, as well as E435Q, M1004I, R88Q, and M1043I (Supplementary Fig. S6A), many of which also tended to be significantly enriched in samples containing a SOLAR1m (Supplementary Fig. S6B and S6C). This class of OTHERm may be appearing later in tumor evolution as secondary mutations that enhance PIK3CA activation, in cis with SOLAR1m (14), and possibly as an acquired resistance mechanism to ET.
In contrast, the mutations that tended to be truncal (N345K, G1049R, indels in the p85 binding and C2 domains, and exon 20 extensions; Supplementary Fig. S6A) were also among the most likely to be detected in samples with no SOLAR1m (Supplementary Fig. S6B and S6C), suggesting a functional redundancy, and that these mutations are likely acting as strong oncogenic drivers in their own right.
Mutational landscape of tumors with OTHERm versus SOLAR1m
We compared the genomic landscape of 11,767 tissue biopsies with PIK3CA mutations and 22,210 tissue biopsies without PIK3CA mutations to study associations of alterations in other genes. (Fig. 2). Genes that were significantly enriched for coalteration with PIK3CA included MAP3K1, CDH1, and TBX3, consistent with previous reports (2, 15, 16). Losses of CDH1 and TBX3 are hallmarks of invasive lobular carcinoma (ILC), while MAP3K1 loss is associated with a luminal A phenotype; both ILC and luminal A breast cancer have higher prevalence of PIK3CAm. Alterations in several components of the PI3K signaling pathway tended to be mutually exclusive with PIK3CA activation: PIK3R1, AKT1, PTEN, and IGF1R, consistent with previous reports (7, 17). Other mutually exclusive alterations were found in BRCA1/2, KDM5A, CCNE1, MCL1, MYC, TP53, LYN, GATA3, CDKN2A/B, RB1, FGFR1, and ZNF703.
The patterns of comutation and mutual exclusivity were strikingly similar among the samples with SOLAR1m (N = 9,467), the samples with SOLAR1m in exon 9 (N = 5,061) and exon 20 (N = 4,287), the samples with OTHERm and no SOLAR1m (N = 2,300) and the subset of those samples with the most common OTHERm, N345K (N = 667; Fig. 2). The overall similarity of the genomic landscape between samples with SOLAR1m and samples with only OTHERm suggests that these different types of PIK3CA mutations occur in similar genomic contexts in ABC.
Benefit of alpelisib in HR+/HER2− patients with PIK3CA mutations
We sought to validate the SOLAR-1 trial results with a real-world dataset in patients with SOLAR1m, as well as explore outcomes for patients with OTHERm who would not have been enrolled in the trial. In patients with available clinical characteristics and outcomes, 163 patients who received ALP+FUL were compared with 134 patients who received FUL alone as treatment for ABC in the second line or later. All of these patients had confirmed HR+/HER2− receptor subtype prior to receiving therapy, and at least one PIK3CA mutation (CONSORT diagram in Supplementary Fig. S2A). The two cohorts were generally well balanced: patient characteristics are shown in Table 1, and variables included in IPTW and achieved balance are shown in Supplementary Fig. S7). Patients in the ALP+FUL arm had higher Eastern Cooperative Oncology Group (ECOG) performance scores prior to the selected line of treatment, were more likely to have had prior CDK4/6 inhibitor therapy, and were treated in later lines. Imbalances between the two groups such as start date range (2012 for FUL vs. May 2019 for ALP+FUL) and number of death events are attributable to the much more recent FDA approval of ALP (2019) than FUL (2002).
Table 1.
Clinical, demographic, and genomic data of the patients in the compared cohorts.
| Characteristic | Alpelisib, fulvestrant (N = 212) | Fulvestrant (N = 155) | FDR |
|---|---|---|---|
| Age at Dx, yrs, Median (IQR) | 56.0 (49.0–64.0) | 56.0 (49.0–65.0) | 0.844 |
| Race | 0.160 | ||
| - Asian | ≤5 | ≤5 | |
| - Black | 17 (8.0%) | 10 (6.5%) | |
| - White | 134 (63.2%) | 113 (72.9%) | |
| - Other race | 40 (18.9%) | 28 (18.1%) | |
| - Not documented | 16 (7.5%) | ≤5 | |
| Stage at Dx | 0.365 | ||
| - I | 35 (16.5%) | 18 (11.6%) | |
| - II | 77 (36.3%) | 50 (32.3%) | |
| - III | 35 (16.5%) | 28 (18.1%) | |
| - IV | 53 (25.0%) | 43 (27.7%) | |
| - IV | 53 (25.0%) | 43 (27.7%) | |
| Metastases sites | 0.921 | ||
| - Bone-only | 28 (13.2%) | 19 (12.3%) | |
| - CNS | 38 (17.9%) | 30 (19.4%) | |
| - Visceral | 146 (68.9%) | 106 (68.4%) | |
| MFI, mo, Median (IQR) | 51.0 (2.8–114.7) | 41.6 (0.0–91.4) | 0.254 |
| Community practice | 188 (88.7%) | 142 (91.6%) | 0.428 |
| Solid biopsy | 174 (82.1%) | 137 (88.4%) | 0.193 |
| Had SOLAR1m | 176 (83.0%) | 117 (75.4%) | 0.285 |
| PTEN loss | 26 (12.3%) | 12 (7.7%) | 0.254 |
| TMB, muts/Mb, Median (IQR) | 2.5 (1.3–5.0) | 2.6 (1.3–6.1) | 0.254 |
| MSI | 0.492 | ||
| - MSI-H | ≤5 | ≤5 | |
| - MSI-I | ≤5 | ≤5 | |
| - MSS | 148 (69.8%) | 115 (74.2%) | |
| - Not documented | 61 (28.8%) | 40 (25.8%) | |
| PD-L1 status | 0.008 a | ||
| - NEGATIVE | 58 (27.4%) | 28 (18.1%) | |
| - POSITIVE | 12 (5.7%) | ≤5 | |
| - Not documented | 142 (67.0%) | 126 (81.3%) | |
| ECOG | 0.026 a | ||
| - 0–1 | 148 (69.8%) | 84 (54.2%) | |
| - 2+ | 19 (9.0%) | 23 (14.8%) | |
| - Not documented | 45 (21.2%) | 48 (31.0%) | |
| Line number | 0.001 a | ||
| - 2 | 50 (23.6%) | 62 (40.0%) | |
| - 3 | 52 (24.5%) | 44 (28.4%) | |
| - 4+ | 110 (51.9%) | 49 (31.6%) | |
| Prior chemotherapy | 127 (59.9%) | 76 (49.0%) | 0.099 |
| Prior CDK4/6i | 206 (97.2%) | 75 (48.4%) | <0.001 a |
| Start date, Median (Range) | 2020-04-18 | 2017-08-22 | <0.001 a |
| (May 20, 2019 to September 28, 2021) | (May 4, 2012 to September 17, 2021) | ||
| Death events | 90 (42.5%) | 115 (74.2%) | <0.001 a |
Note: Because alpelisib was approved in 2019, the average date of alpelisib/fulvestrant therapy line start is about 3 years later than the fulvestrant-alone cohort. Patients tended to be treated with alpelisib in a later line (>50% in line 4+ vs. 32% for fulvestrant) and nearly all (97%) had been treated with a CDK4/6 inhibitor previously. Patients treated with alpelisib had a higher level of functioning prior to the selected line of treatment, and higher rate of PD-L1 positivity.
Abbreviations: CDK4/6, CDK4/6 inhibitor; Dx, diagnosis; ECOG, Eastern Cooperative Oncology Group performance score; IQR, interquartile range; MFI, metastasis-free interval; MSI: microsatellite instability; mut/Mb, mutations per megabase; TMB, tumor mutational burden.
aFDR < 0.05.
Compared with the PIK3CAm cohort in the SOLAR-1 trial, the patients in this real-world cohort had younger median age (56 vs. 63–64), higher ECOG scores (9% and 15% of ALP+FUL and FUL patients had scores of 2+ whereas SOLAR-1 was limited to 0–1), and higher rates of visceral and central nervous system (CNS) metastases (SOLAR-1 excluded patients with CNS metastases). In the SOLAR-1 cohort, 5% and 6% of patients on the ALP+FUL and FUL arms had been treated with cyclin-dependent kinase 4/6 inhibitors (CDK4/6i), compared with 97% of ALP+FUL and 48% of FUL arm patients in the real-world cohort.
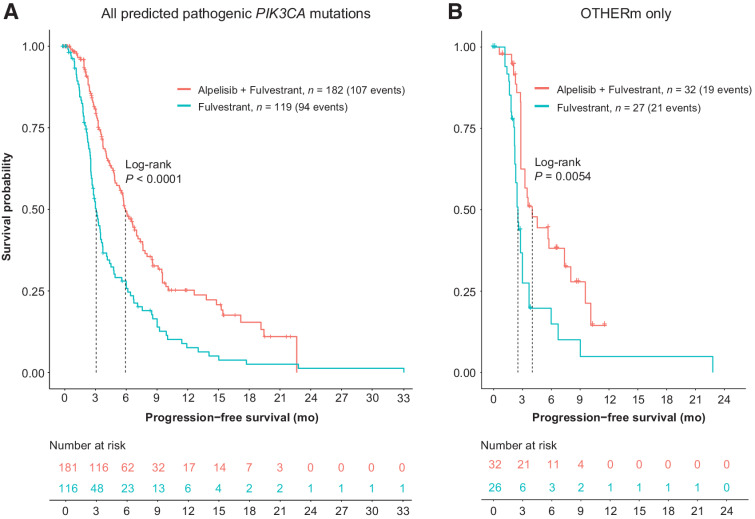
Despite ALP+FUL often being used in a later-line setting than FUL, patients with PIK3CA mutations had longer rwPFS with ALP: median rwPFS was 5.9 months on ALP+FUL (95% CI: 5.1–7.4) versus 3.1 months on FUL (95% CI: 2.7–3.7), log-rank P < 0.0001 (Fig. 3). Patients receiving alpelisib also had longer rwOS: 21.4 months (95% CI: 14.1–24.5) after ALP+FUL regimen start versus 5.3 months (95% CI: 5.3–11.7) after FUL line start, log-rank P < 0.0001 (Supplementary Fig. S8).
Figure 3.
PFS among patients with PIK3CA-mutated HR+HER2− breast cancer was significantly longer on alpelisib+fulvestrant than fulvestrant alone. A, A total of 301 patients who had documented progression record were included in comparison. Kaplan–Meier curves represent PFS estimation based on real PFS adjusted using IPTW method (see Supplementary Fig. S7 for further details on IPTW). B, Subset analysis. In patients with breast cancer harboring OTHERm and no SOLAR1m, alpelisib treatment resulted in significantly longer PFS than fulvestrant alone.
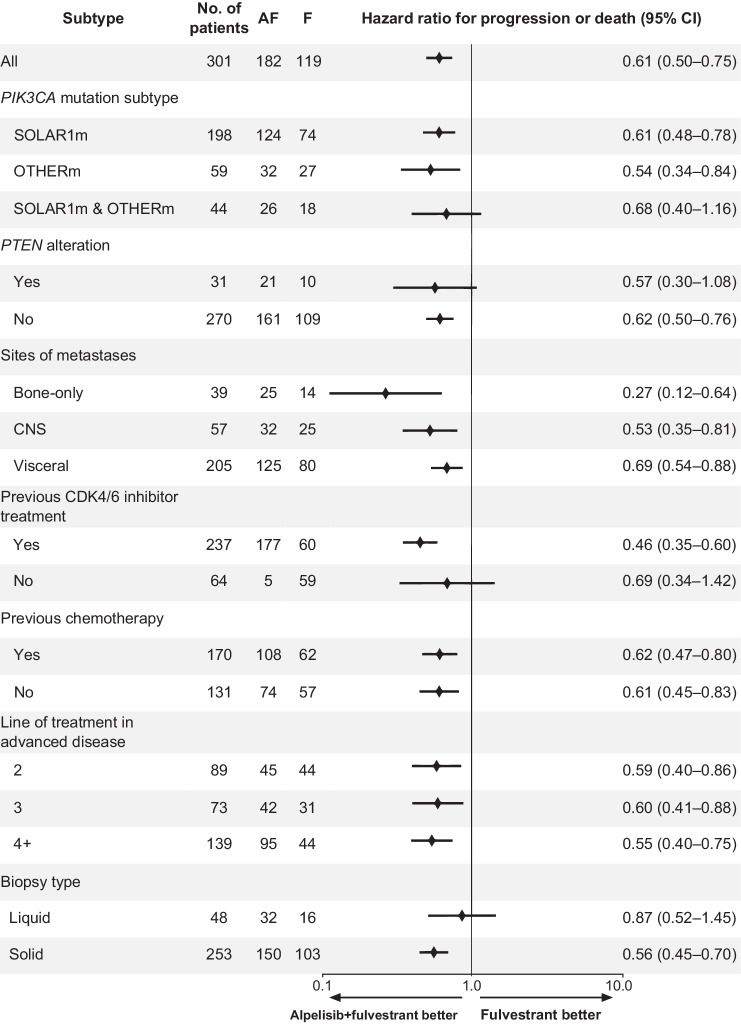
The majority of patients in this analysis had a SOLAR1m (Table 1). We examined the patient subsets with and without SOLAR1m separately (Fig. 3B; Supplementary Fig. S9). In the 59 patients whose tumors harbored OTHERm and no SOLAR1m, median rwPFS was 4 months on ALP+FUL (95% CI: 2.8–10.1) versus 2.5 months on FUL (95% CI: 2.2–3.7), log-rank P = 0.0054 (Fig. 3B). Subgroup analysis of PFS showed consistent benefit of treatment with ALP+FUL over FUL across the selected subgroups (Fig. 4), including similar HRs for patients with SOLAR1m (N = 198), OTHERm (N = 59), or both (N = 44).
Figure 4.
Subgroup analysis of PFS in the HR+/HER2−PIK3CA-mutated breast cancer cohort. Alpelisib showed consistent benefit of treatment over fulvestrant across the selected subgroups. Confidence intervals have not been adjusted for multiple comparisons. Some patients had more than one PIK3CA mutation. AF, alpelisib/fulvestrant combination therapy; F, fulvestrant monotherapy; CNS, central nervous system; CDK4/6 inhibitor, abemaciclib, palbociclib, or ribociclib.
PFS on alpelisib in patients with OTHERm and no SOLAR1m
The set of patients with OTHERm had a heterogeneous collection of mutations. We examined whether certain mutations or mutations in certain domains were more predictive of benefit. The swimmer plot in Fig. 5 depicts the rwPFS of 77 patients (including the 59 considered in the previous analysis) with OTHERm but no SOLAR1m who were treated with ALP in any combination, at any point in their treatment course, and regardless of hormone receptor subtype. Consecutive treatment lines that contained ALP in different combinations are presented contiguously in this analysis. Twenty nine patients (38%) were treated with ALP for longer than 6 months, including 17 still on therapy at the analysis cut-off date. Fourteen patients (18%) had been on therapy less than 6 months without a progression event at analysis cutoff.
Figure 5.
PFS among patients with OTHERm treated with alpelisib in any combination. Swimmer plot representing rwPFS of 77 patients with OTHERm and no SOLAR1m, clustered by exon location of the OTHERm. For patients who had OTHERm in more than one exon, swimmer lanes are presented in all relevant exons. The column labeled “#” represents patient identification number, in order of descending PFS. Treatment line number (“ALP line”): whether a patient had a tissue or liquid biopsy profiled, receptor subtype, and prior chemotherapy and CDK4/6 inhibitor treatment are indicated for each patient.
The PIK3CA mutations in this cohort were diverse, with no discernable association between specific variants and/or protein domains and longer rwPFS. The 29 patients with rwPFS longer than 6 months harbored 37 mutations in: N345K (exon 4; 10), the p85 binding domain (exon 1; 9), the C-terminal tail (exon 20; 9), the C2 domain (exon 7; 4), E726K (exon 13; 3), and helical domain (exon 9; 2; Fig. 5). In 8 of these 29 patients, a liquid biopsy had been used to detect the OTHERm.
Discussion
To our knowledge, this is the first study to examine survival outcomes on ALP in the population with PIK3CA mutations known to be oncogenic through preclinical data (7, 18–21) but not included in the list of mutations examined in the SOLAR-1 trial. This study validates SOLAR-1 findings using a real-world cohort. It also shows that a sizeable portion (20%) of patients with PIK3CA mutations would not be identified by the companion diagnostic PCR hotspot test. Crucially, this study finds that these patients with diverse OTHERm also have longer rwPFS on ALP+FUL compared with FUL alone.
Our study suggests that 20% of PIK3CA-mutated breast cancer would have been missed by therascreen companion diagnostic PCR testing. When comparing NGS-based CGP with PCR hotspot testing in a cohort of 128 patients with results from both assays, CGP testing did not miss any samples with mutations detected by PCR, but PCR testing missed some samples detected by CGP (including mutations the PCR test was designed to detect). It is worth considering that PCR hotspot testing may be less sensitive than CGP, and that CGP can simultaneously yield information about other genomic alterations relevant to breast cancer (germline and somatic BRCA1/2 alterations, PTEN loss, ESR1 mutations associated with ET resistance, RB1 mutations associated with CDK4/6i resistance) as well as genomic biomarkers such as tumor mutational burden. In the phase II FAKTION trial, the AKT1 inhibitor capivasertib + fulvestrant was initially reported to improve PFS and OS compared with fulvestrant alone in patients with HR+ ABC regardless of alternations in the PI3K pathway using digital-droplet PCR detection of just four mutations in exons 9 and 20 (22). However, when NGS was used, the benefit of combination therapy was only seen in patients with alterations in PIK3CA, AKT1, and PTEN (23), confirming the importance of CGP in understanding the benefit of targeted agents.
Vasan and colleagues (ref. 14; PMID: 31699932) reported enhanced sensitivity to alpelisib and GDC-077 in cell lines engineered with double-mutated PIK3CA over mutations E545K or H0147R alone, and greater overall rate of response to taselisib in patients with two mutations in a retrospective analysis of the SANDPIPER trial. We did not observe this trend in our cohort. In 44 patients who had both SOLAR1m and OTHERm, the observed HR (0.68) was similar to that seen in patients with SOLAR1m (0.61) or OTHERm (0.54) alone.
This study corroborates previous findings that the landscape of genomic alterations in breast cancer with PIK3CA mutations differs from PIK3CA wild-type breast cancer, with characteristic enrichments in MAP3K1, CDH1, and TBX3 loss-of-function mutations, low rates of coalteration in PIK3R1, AKT1, IGF1R, and PTEN (likely due to redundancies in PI3K signaling), and significantly lower rates of BRCA1/2, KDM5A, and MYC alterations. These patterns were found to be highly similar across samples with SOLAR1m and with OTHERm.
Our analysis of liquid biopsies from patients with breast cancer showed that 94% of samples had detectable ctDNA, and highlights the value of reporting an estimate of circulating TF in a liquid biopsy. When TF is high (as it was in 37% of our paired cohort), liquid biopsy has excellent NPV and the currently National Comprehensive Cancer Network recommended reflex to tissue for a negative liquid biopsy may have less value in these sample. On the other hand, when TF is low, detection of PIK3CA mutations is still possible, but a negative result, particularly with no other driver mutations detected, cannot be taken at face value and should be confirmed with a tissue biopsy. As an interesting aside, in the subgroup analysis of rwPFS in the HR+/HER2− cohort, the patients with a liquid biopsy result appeared to have less significant benefit from ALP+FUL (Fig. 4). We speculate that presence of ctDNA may be an unfavorable prognostic factor that erodes some of the benefit of a targeted therapy. Nevertheless, as can be seen in the OTHERm cohort swimmer plot, many patients who had a PIK3CA mutation detected by liquid biopsy continued on ALP therapy for more than 6 months. Liquid biopsy presents an attractive, less invasive option for genomic profiling when metastatic tissue is hard to obtain and will tend to yield informative results in patients with the most urgent need—those with aggressive, high-ctDNA-shedding disease.
A limitation of this study is that it is a retrospective analysis, where the two treatment arms being compared had different median start dates. Given the larger differential in rwOS than rwPFS observed, it is possible that patients on the ALP+FUL arm had better prognosis than those on the FUL arm. Because this is not a randomized controlled trial, it is also difficult to exclude the possibility of systematic differences between patients on the two treatment arms. It is also likely that changes in standards of care over that time affect the analysis (e.g., the widespread use of CDK4/6i). Survival in this population has increased over time, presumably due to more effective treatment options.
Although a recent SOLAR-1 analysis reported no significant difference in health-related quality of life between the ALP and placebo arms of the trial (24), ALP does have a known side-effects and adverse events profile. It is thus imperative to establish whether patients with the rarer varieties of PIK3CA mutations have clinically meaningful PFS benefit. Prospective trials or larger retrospective studies are needed to assess this question.
There are several other outstanding questions on which prospective trials could shed light. One question is whether PI3K inhibitors could be effective for tumors with other PI3K pathway component alterations such as PTEN and PIK3R1 loss and AKT1 activation. A related question is whether these alterations as comutations in PIK3CAm tumors have a modifying effect on alpelisib response. It is worth noting a recent report that found baseline phosphoprotein levels as functional biomarkers predict benefit from PI3K pathway inhibitors (25), although much work remains to validate and commercially scale such assays to reach the companion diagnostic standards of PCR and NGS. Alpelisib is being evaluated in the treatment of HER2+ ABC, where PI3K signaling may be a form of resistance to HER2 inhibition (26). The patient with OTHERm who stayed on alpelisib therapy the longest in our clinically annotated cohort had HR+/HER2+ ABC and was being treated with alpelisib, fulvestrant, and trastuzumab in combination. Ongoing studies are evaluating the efficacy of alpelisib in HER2+ and in triple-negative breast cancers that harbor PIK3CA mutations. Finally, with the advent of more selective, H1047R-specific inhibitors of PI3Ka like LOX-22783 (27) or inhibitors targeting downstream targets like AKT1 (23), it will be important to compare the efficacy and durability of response to ATP-competitive inhibitors like ALP.
Supplementary Material
Supplementary Figure 1. CONSORT diagram of genomic analyses cohorts
Supplementary Figure 2. CONSORT diagrams for the cohorts with clinicogenomic data
Supplementary Figure 3. PIK3CA variants detected in paired tissue and liquid samples.
Supplementary Figure 4. Impact of ctDNA fraction on sensitivity of detection of PIK3CAmutations.
Supplementary Figure 5. Comparison of performance of comprehensive genomicprofiling by next-generation sequencing (NGS) and PCR hotspot testing.
Supplementary Figure 6. OTHERm variants show a range of subclonality andindependent oncogenicity
Supplementary Figure 7. Individual covariate balance before and after inverse probabilityof treatment weighting.
Supplementary Figure 8. Overall survival among patients with PIK3CA-mutatedHR+HER2- breast cancer treated with alpelisib/fulvestrant versus fulvestrant alone.
Supplementary Figure 9. Progression-free survival (PFS) among patients with SOLAR1mtreated with alpelisib+fulvestrant versus fulvestrant monotherapy.
Acknowledgments
We thank the patients, families, and physicians who ordered comprehensive genomic profiling from Foundation Medicine Inc. We thank James Creeden, Jeffrey Venstrom, Mason lsrael, and Kimberly McGregor for their support in the early phase of the manuscript.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Disclosures
H.S. Rugo reports grants from Pfizer, Novartis, Eli Lilly, Genentech/Roche, OBI, Merck, Gilead Sciences, Daiichi Sankyo, Seattle Genetics, Sermonix, AstraZeneca, Astellas, Veru, GSK, Taiho, AMBRX, and Pionyr; nonfinancial support from Merck, AstraZeneca, and Gilead; and personal fees from PUMA, NAPO, and Blueprint outside the submitted work. K. Raskina reports personal fees from Foundation Medicine and other support from Roche during the conduct of the study. A.B. Schrock reports other support from Foundation Medicine and Roche during the conduct of the study. R.W. Madison reports personal fees from Foundation Medicine during the conduct of the study as well as other support from Roche Holdings AG outside the submitted work. R.P. Graf reports personal fees from Foundation Medicine during the conduct of the study as well as personal fees from Epic Sciences outside the submitted work. E.S. Sokol reports other support from Foundation Medicine and Roche during the conduct of the study. S. Sivakumar reports personal fees from Foundation Medicine and other support from Roche during the conduct of the study. J.K. Lee reports personal fees from Foundation Medicine and Roche during the conduct of the study. V. Fisher reports grants from Roche outside the submitted work. G.R. Oxnard reports personal fees from Foundation Medicine and Roche during the conduct of the study. H. Tukachinsky reports personal fees from Foundation Medicine and other support from Roche during the conduct of the study. No other disclosures were reported.
Authors' Contributions
H.S. Rugo: Conceptualization, writing–review and editing. K. Raskina: Formal analysis, writing–original draft, writing–review and editing. A.B. Schrock: Supervision, writing–review and editing. R.W. Madison: Writing–review and editing. R.P. Graf: Writing–review and editing. E.S. Sokol: Writing–review and editing. S. Sivakumar: Writing–review and editing. J.K. Lee: Methodology, writing–review and editing. V. Fisher: Methodology, writing–review and editing. G.R. Oxnard: Supervision, writing–review and editing. H. Tukachinsky: Conceptualization, formal analysis, writing–original draft, writing–review and editing.
References
- 1. Vasan N, Cantley LC. At a crossroads: how to translate the roles of PI3K in oncogenic and metabolic signalling into improvements in cancer therapy. Nat Rev Clin Oncol 2022;19:471–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mosele F, Stefanovska B, Lusque A, Tran Dien A, Garberis I, Droin N, et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann Oncol 2020;31:377–86. [DOI] [PubMed] [Google Scholar]
- 3. Dent S, Cortes J, Im YH, Dieras V, Harbeck N, Krop IE, et al. Phase III randomized study of taselisib or placebo with fulvestrant in estrogen receptor-positive, PIK3CA-mutant, HER2-negative, advanced breast cancer: the SANDPIPER trial. Ann Oncol 2021;32:197–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med 2019;380:1929–40. [DOI] [PubMed] [Google Scholar]
- 5. Andre F, Ciruelos EM, Juric D, Loibl S, Campone M, Mayer IA, et al. Alpelisib plus fulvestrant for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: final overall survival results from SOLAR-1. Ann Oncol 2021;32:208–17. [DOI] [PubMed] [Google Scholar]
- 6. Martinez-Saez O, Chic N, Pascual T, Adamo B, Vidal M, Gonzalez-Farre B, et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res 2020;22:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen L, Yang L, Yao L, Kuang XY, Zuo WJ, Li S, et al. Characterization of PIK3CA and PIK3R1 somatic mutations in Chinese breast cancer patients. Nat Commun 2018;9:1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khoury K, Tan AR, Elliott A, Xiu J, Gatalica Z, Heeke AL, et al. Prevalence of phosphatidylinositol-3-Kinase (PI3K) pathway alterations and co-alteration of other molecular markers in breast cancer. Front Oncol 2020;10:1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 2013;31:1023–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Milbury CA, Creeden J, Yip WK, Smith DL, Pattani V, Maxwell K, et al. Clinical and analytical validation of FoundationOne(R)CDx, a comprehensive genomic profiling assay for solid tumors. PLoS One 2022;17:e0264138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Woodhouse R, Li M, Hughes J, Delfosse D, Skoletsky J, Ma P, et al. Clinical and analytical validation of FoundationOne Liquid CDx, a novel 324-Gene cfDNA-based comprehensive genomic profiling assay for cancers of solid tumor origin. PLoS One 2020;15:e0237802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spangle JM, Von T, Pavlick DC, Khotimsky A, Zhao JJ, Roberts TM. PIK3CA C-terminal frameshift mutations are novel oncogenic events that sensitize tumors to PI3K-alpha inhibition. Proc Natl Acad Sci U S A 2020;117:24427–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Q therascreen. PIK3CA RGQ PCR Kit Instructions for Use (Handbook). Available from:https://www.accessdata.fda.gov/cdrh_docs/pdf19/P190001C.pdf.
- 14. Vasan N, Razavi P, Johnson JL, Shao H, Shah H, Antoine A, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kalpha inhibitors. Science 2019;366:714–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015;163:506–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nixon MJ, Formisano L, Mayer IA, Estrada MV, Gonzalez-Ericsson PI, Isakoff SJ, et al. PIK3CA and MAP3K1 alterations imply luminal A status and are associated with clinical benefit from pan-PI3K inhibitor buparlisib and letrozole in ER+ metastatic breast cancer. NPJ Breast Cancer 2019;5:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, Neve RM, Kuo WL, Davies M, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res 2008;68:6084–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dogruluk T, Tsang YH, Espitia M, Chen F, Chen T, Chong Z, et al. Identification of variant-specific functions of PIK3CA by rapid phenotyping of rare mutations. Cancer Res 2015;75:5341–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gymnopoulos M, Elsliger MA, Vogt PK. Rare cancer-specific mutations in PIK3CA show gain of function. Proc Natl Acad Sci U S A 2007;104:5569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jin N, Keam B, Cho J, Lee MJ, Kim HR, Torosyan H, et al. Therapeutic implications of activating noncanonical PIK3CA mutations in head and neck squamous cell carcinoma. J Clin Invest 2021;131:e150335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ng PK, Li J, Jeong KJ, Shao S, Chen H, Tsang YH, et al. Systematic functional annotation of somatic mutations in cancer. Cancer Cell 2018;33:450–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jones RH, Casbard A, Carucci M, Cox C, Butler R, Alchami F, et al. Fulvestrant plus capivasertib versus placebo after relapse or progression on an aromatase inhibitor in metastatic, oestrogen receptor-positive breast cancer (FAKTION): a multicentre, randomised, controlled, phase 2 trial. Lancet Oncol 2020;21:345–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones RH, Casbard AC, Carucci M, Ingarfield-Herbert K, Butler R, Morgan S, et al. Fulvestrant plus capivasertib versus fulvestrant plus placebo after relapse or progression on an aromatase inhibitor in metastatic, estrogen receptor–positive breast cancer (FAKTION): overall survival and updated progression-free survival data with enhanced biomarker analysis. J Clin Oncol 40: 16s, 2022(suppl; abstr 1005). [Google Scholar]
- 24. Ciruelos EM, Rugo HS, Mayer IA, Levy C, Forget F, Delgado Mingorance JI, et al. Patient-reported outcomes in patients with PIK3CA-mutated hormone receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer from SOLAR-1. J Clin Oncol 2021;39:2005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shi Z, Wulfkuhle J, Nowicka M, Gallagher RI, Saura C, Nuciforo PG, et al. Functional mapping of AKT signaling and biomarkers of response from the FAIRLANE trial of Neoadjuvant Ipatasertib plus Paclitaxel for triple-negative breast cancer. Clin Cancer Res 2022;28:993–1003, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rasti AR, Guimaraes-Young A, Datko F, Borges VF, Aisner DL, Shagisultanova E. PIK3CA mutations drive therapeutic resistance in human epidermal growth factor receptor 2-positive breast cancer. JCO Precis Oncol 2022;6:e2100370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klippel A, Wang R, Puca L, Faber AL, Shen W, Bhagwat SV, et al. Preclinical characterization of LOX-22783, a highly potent, mutant-selective and brain-penetrant allosteric PI3Kα H1047R inhibitor [abstract]. In:Proceedings of the AACR-NCI-EORTC Virtual International Conference on Molecular Targets and Cancer Therapeutics; 2021 Oct 7–10. Philadelphia (PA): AACR; Mol Cancer Ther 2021;20(12 Suppl):Abstract nr P142. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. CONSORT diagram of genomic analyses cohorts
Supplementary Figure 2. CONSORT diagrams for the cohorts with clinicogenomic data
Supplementary Figure 3. PIK3CA variants detected in paired tissue and liquid samples.
Supplementary Figure 4. Impact of ctDNA fraction on sensitivity of detection of PIK3CAmutations.
Supplementary Figure 5. Comparison of performance of comprehensive genomicprofiling by next-generation sequencing (NGS) and PCR hotspot testing.
Supplementary Figure 6. OTHERm variants show a range of subclonality andindependent oncogenicity
Supplementary Figure 7. Individual covariate balance before and after inverse probabilityof treatment weighting.
Supplementary Figure 8. Overall survival among patients with PIK3CA-mutatedHR+HER2- breast cancer treated with alpelisib/fulvestrant versus fulvestrant alone.
Supplementary Figure 9. Progression-free survival (PFS) among patients with SOLAR1mtreated with alpelisib+fulvestrant versus fulvestrant monotherapy.
Data Availability Statement
The data supporting the findings of this study originated from Foundation Medicine, Inc and Flatiron Health, Inc. These deidentified data may be made available upon request, and are subject to a license agreement with FH and FMI; interested researchers should contact the corresponding author and cgdb-fmi@flatiron.com to determine licensing terms.