Abstract
The phospholipid composition of Hydrogenobacter thermophilus strain TK-6, an obligately chemolithoautotrophic, extremely thermophilic hydrogen bacterium, was analyzed. Two of four phospholipids detected from the strain were assumed to be phosphatidylinositol and phosphatidylglycerol. An aminophospholipid named PX, whose content among the phospholipids was 65%, was found to have a novel chemical structure by analysis of the dilyso form with nuclear magnetic resonance and fast atom bombardment-mass spectrometry (FAB-MS) and by analysis of the intact PX with FAB-MS as 1,2-diacyl-3-O-(phospho-2′-O-(1′-amino)-2′,3′,4′,5′-pentanetetrol)-sn-glycerol. Structurally similar phospholipids have been identified in Methanospirillum hungatei, Methanolacinia paynteri, and Methanogenium cariaci, which all belong to the Archaea.
Hydrogenobacter thermophilus strain TK-6 is an obligately chemolithoautotrophic, extremely thermophilic hydrogen bacterium whose optimal growth temperature is around 70 to 75°C (7). Phylogenetic analysis based on the 16S ribosomal DNA sequence showed that the strain has the deepest branching point among the Bacteria (9). In accordance with this, distinctive characteristics, such as unusual composition of cellular fatty acids (C18:0 and C20:1 as major fatty acids) and a novel sulfur-containing quinone called methionaquinone, have been reported so far (5–7). In this paper, we report the phospholipid composition of TK-6 and also the chemical structure of a new aminophospholipid from this strain.
MATERIALS AND METHODS
Strain used in this study.
H. thermophilus strain TK-6 (DSM 6534, IAM 12695) was used in this study (7). The strain was cultivated under chemolithoautotrophic conditions with H2 gas as an energy source and O2 gas as an electron acceptor, as described elsewhere (7).
Phospholipid analysis.
Lipids were extracted from lyophilized cells by the Bligh-Dyer method (1). A silica gel 60 F254 thin-layer chromatography (TLC) plate (20 by 20 cm, 0.25 mm thick; Merck) was used for the analysis of phospholipids. Development was performed with the solvent chloroform-methanol-water (65:25:4). Detection was performed with either the Dittmer reagent for phospholipids (2) or the ninhydrin reagent for amino groups. In a separate experiment, the extracted lipids were applied in a line onto the TLC plate. After development, both edges of the plate were cut to give two strips with a width of 1 cm and the strips were sprayed with Dittmer reagent. Then, the regions corresponding to the phospholipids PW, PX, PY, and PZ (see “Phospholipid composition” below) were scraped and extracted with solvent (chloroform-methanol, 2:1). After evaporation, weight was measured and used for quantitation.
Purification of PX.
Total lipids were extracted with the solvent (chloroform-methanol, 2:1) from the wet cells (261 g). After the extract was dried, almost all the quinone (methionaquinone) was removed by acetone extraction. Then, PX was purified by following the procedure described above except that preparative TLC plates (silica gel 60 F254 TLC plate [20 by 20 cm, 2 mm thick, Merck]) were used.
Mild alkaline hydrolysis of PX.
Purified PX (330 mg) was dissolved in a solvent consisting of 4.0 ml of CHCl3, 37.5 ml of C2H5OH, 3.25 ml of pure water, and 1.25 ml of 1 M NaOH, and the solution was incubated for 30 min at 37°C. Two milliliters of ethyl formate was poured into the reaction mixture to stop the reaction, and the total solution was evaporated. Then, 5 ml of water, 3.35 ml of 2-butanol, and 6.65 ml of CHCl3 were added, followed by extraction and centrifugation for 30 min at 185 × g. The upper layer, which contained the PX dilyso form, was evaporated, lyophilized, and dissolved in 2 ml of pure water.
Purification of the PX dilyso form.
The above-described solution containing the PX dilyso form was injected in 20-μl aliquots into a high-pressure liquid chromatography system (D-6500; Hitachi). High-pressure liquid chromatography was carried out under the following conditions: column, Shim-pack CLC-SIL(M), 4.6 mm (diameter) by 25 cm (Shimadzu); column temperature, 45°C; solvent, hexane–2-propanol-water (38:51:10); and flow rate, 1 ml/min. Fractionation was performed every minute, and each small portion was applied to TLC analysis to detect the PX dilyso form by the ninhydrin reaction. The 32- to 47-min fractions were collected, evaporated, and lyophilized. The lyophilized compound (4.7 mg) was used for the structural analysis described below.
Analysis of the PX dilyso form and PX.
Structural analyses of the PX dilyso form were performed by nuclear magnetic resonance (NMR) and high-resolution fast atom bombardment-mass spectrometry (HR-FAB-MS). Also, structural analysis of PX was done by FAB-MS. NMR spectra were collected on a JEOL JMN-A500 spectrometer with D2O as the solvent and acetone as an external standard. The measurements of 1H and 13C were performed at 500 and 125 MHz, respectively. The chemical shifts (δ) are reported in parts per million, and the coupling constants (J values) are in hertz. NMR experiments included 1H-1H COSY (correlation spectroscopy), HMQC (heteronuclear multiple quantum coherence), and HMBC (heteronuclear multiple-bond correlation spectroscopy). FAB-MS (positive or negative mode) and HR-FAB-MS (positive mode) was carried out on a JEOL JMS-SX102 spectrometer using a glycerol matrix.
RESULTS
Phospholipid composition.
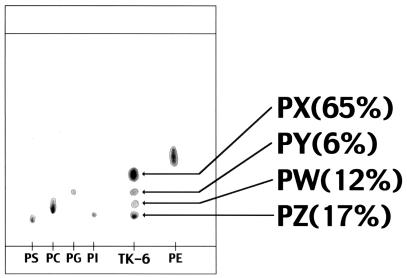
Extracted lipids were developed on a silica gel TLC, followed by color development with the Dittmer reagent. Four phospholipids, named PX, PY, PW, and PZ, were detected (Fig. 1). PX also gave a red-purple color with the ninhydrin reagent, showing that it is an aminophospholipid. PY and PZ were thought to be phosphatidylglycerol and phosphatidylinositol, respectively. Since the major phospholipid (PX), which comprised about 40% of lipids extracted by the Bligh-Dyer method, and the minor phospholipid (PW) could not be identified, structural analysis of PX was performed. However, due to low yield, further structural analysis of PW was not possible.
FIG. 1.
TLC analysis of phospholipids from H. thermophilus strain TK-6. The percentages show weight distribution among four phospholipids detected in the strain. Abbreviations: PS, phosphatidylserine; PC, phosphatidylcholine; PG, phosphatidylglycerol; PI, phosphatidylinositol; PE, phosphatidylethanolamine.
Purification and structural analysis of PX.
Lipids were extracted from wet cells (261 g), and PX (330 mg) was purified by preparative silica gel TLC. Mild alkaline hydrolysis of PX gave undecomposed PX, the PX monolyso form, and the PX dilyso form. Further hydrolysis of the isolated PX monolyso form yielded the PX dilyso form. These facts together with the fact that PX was digested with phospholipase A2 (data not shown) showed that PX is an acyl-type phospholipid with two acyl chains within a molecule.
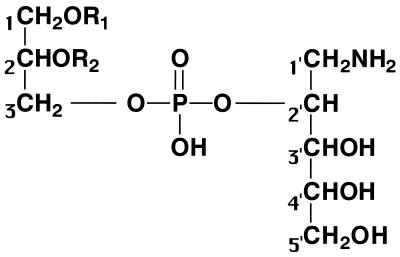
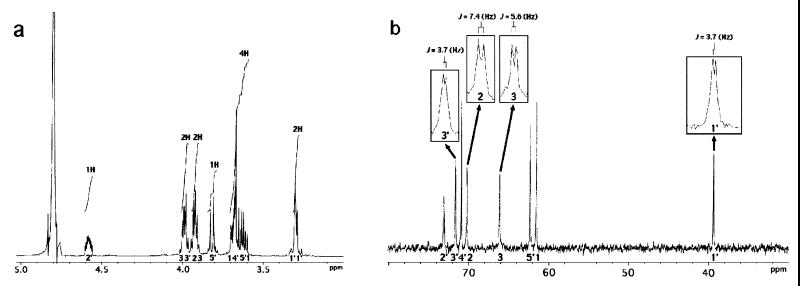
The molecular formula of the dilyso form (referred to here as structure 1) (Fig. 2) was determined to be C8H20O9NP by its HR-FAB-MS spectrum. By analysis of 1H-NMR (Fig. 3a), 13C-NMR (Fig. 3b), 1H-1H COSY, HMQC, and HMBC spectra of structure 1, two partial structures, 1CH2(O—)CH(O—) CH2(O—) and 1′CH2(N—)CH(O—)CH(O—)CH(O—) CH2(O—), which contained all the carbon atoms in structure 1, were identified. In the 13C NMR spectrum of structure 1, the signals of C2, C3, C1′, and C3′ were split into doublets by the carbon-phosphorus couplings with 2JC,P or 3JC,P values of 7.4, 5.6, 3.7, and 3.7 Hz, respectively. This observation and the number of phosphorus atoms in structure 1 showed that both C3 and C2′ were phosphorylated to combine the two partial structures. Finally, based on the molecular formula, the dilyso form was determined to correspond to structure 1. Taking into account that PX has two acyl chains within a molecule, the overall structure of PX was assumed to be structure 2 (Fig. 2). Structure 2 was confirmed by analyzing PX through FAB-MS. The positive mode (FAB+), which gives ion peaks of (M + Na)+ species, gave several ion peaks: 858, 872, 886, 900, 912, and 926 (m/z). The negative mode (FAB−), which gives ion peaks of (M − H)− species, gave the following ion peaks: 834, 848, 862, 876, 888, and 902 (m/z). These ion peaks are thought to derive from the PX molecule with the following respective pairs of fatty acids: C16:0 and C20:1, C16:0 and CΔ21:0, C18:0 and C20:1, C18:0 and CΔ21:0, C20:1 and C20:1, and C20:1 and CΔ21:0. Thus, PX is a novel aminophospholipid that had never been reported. The assignment of spectra of structure 1 was as follows: HR-FAB-MS (positive, glycerol matrix) m/z 306.0954 (M + H)+ (calculated for C8H21O9NP, 306.0954); δH (D2O, 500 MHz) 4.58 (m, H-2′), 4.00 (dd, J = 6.5, 12.5 Hz, H-3a), 3.99 (dd, J = 3.5, 8.0 Hz, H-3′), 3.92 (qui, J = 6.0 Hz, H-2), 3.92 (q, J = 6.0 Hz, H-3b), 3.82 (dd, J = 6.0, 12.5 Hz, H-5′a), 3.70 (q, J = 6.5 Hz, H-1a), 3.68 (ddd, J = 3.5, 6.5, 6.5 Hz, H-4′), 3.62 (dd, J = 6.5, 13.5 Hz, H-5′b), 3.60 (dd, J = 6.5, 15.5 Hz, H-1b), 3.31 (dd, J = 3.5, 13.5 Hz, H-1′a), and 3.28 (dd, J = 6.5, 13.5 Hz, H-1′b); δC (D2O, 125 MHz) 73.1 (broad, C-2′), 71.7 (d, J = 3.7 Hz, C-3′), 70.9 (C-4′), 70.2 (d, J = 7.4 Hz, C-2), 66.1 (d, J = 5.6 Hz, C-3), 62.3 (C-5′), 61.6 (C-1), and 39.4 (d, J = 3.7 Hz, C-1′).
FIG. 2.
Chemical structures of PX and the PX dilyso form. In the PX dilyso form (structure 1), R1 and R2 are H; in PX (structure 2), R1 and R2 are acyl chains, not alkyl chains.
FIG. 3.
NMR spectra of the PX dilyso form. (a) 1H-NMR; (b) 13C-NMR.
DISCUSSION
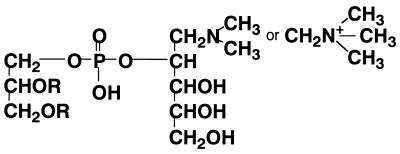
In this paper, we report the phospholipid composition of H. thermophilus strain TK-6, with special emphasis on the chemical structure of a new aminophospholipid. Although the stereochemistry of glycerol moiety of PX is not known, structurally similar phospholipids have been reported for the archaeon Methanospirillum hungatei (Fig. 4) (3, 11). The major difference between the phospholipids of the two strains is that, instead of being an ether-type phospholipid, like that in M. hungatei, the phospholipid from H. thermophilus strain TK-6 is an acyl type. Among Archaea, phospholipids with (methylated) aminopentanetetrol have been detected only in M. hungatei, Methanolacinia paynteri, and Methanogenium cariaci, which all belong to the Methanomicrobiaceae (8). In the phylogenetic tree based on 16S ribosomal DNA sequences, the family Methanomicrobiaceae has a late branching point among Archaea, while the family Aquificaceae, where H. thermophilus strain TK-6 belongs, has the earliest branching point among the Bacteria. Therefore, it will be quite interesting to investigate if phospholipids with (methylated) aminopentanetetrol can be found in other microorganisms, especially among Bacteria.
FIG. 4.
Chemical structure of aminophospholipid from M. hungatei. R represents a saturated, 20-carbon isoprenoid chain in ether linkage to the glycerol backbone.
In H. thermophilus strain TK-6, the major cellular fatty acids are reported to be C18:0 and C20:1 (7). Among six assigned ion peaks of FAB-MS of PX, all except one were shown to contain C18:0 and/or C20:1 as fatty acids. This result clearly shows that C18:0 and C20:1 are also major fatty acids in PX.
Among strains which belong to the family Aquificaceae, the complex lipid analysis has been performed only for Aquifex pyrophilus (4). In A. pyrophilus, an aminophospholipid accounts for 57% of the lipid, while a glycolipid and a phospholipid account for 9.8 and 32% of the lipid, respectively. It is very interesting that PX accounts for 40% of the lipid in H. thermophilus strain TK-6. It has also been reported that in A. pyrophilus (4), the main core lipid (66%) is an alkyl glycerol diether, which is quite different from the case of H. thermophilus strain TK-6. We have not detected any ether-type lipids in TK-6. Therefore, the structure of aminophospholipid from A. pyrophilus should be clarified in future studies.
It would also be interesting to know how the aminophospholipid, especially the aminopentanetetrol part, is biosynthesized in H. thermophilus strain TK-6, where a reductive tricarboxylic acid cycle is operative as the CO2 fixation pathway (10, 12–14). However, very little is known about gluconeogenesis in TK-6, and thus, future experiments should focus on clarification of gluconeogenesis in this strain.
ACKNOWLEDGMENTS
We thank Hirobumi Imai, of the University of Tokyo, for technical assistance.
This work was supported in part by a grant-in-aid from the Ministry of Education, Science, and Culture of Japan and in part by collaboration in a Core University Program between Yamaguchi University and Kasetsart University supported by the Scientific Cooperation Program agreed upon by the Japan Society for Promotion of Science (JSPS) and the National Research Council of Thailand (NRCT).
REFERENCES
- 1.Bligh E G, Dyer W J. A rapid method of total lipid extraction and purification. Can J Biochem. 1959;234:466–468. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 2.Dittmer J D, Lester R L. A simple, specific spray for the detection of phospholipids on thin-layer chromatograms. J Lipid Res. 1964;5:126–127. [PubMed] [Google Scholar]
- 3.Ferrante G, Ekiel I, Sprott G D. Structures of diether lipids of Methanospirillum hungatei containing novel head groups N,N-dimethylamino- and N,N,N-trimethylaminopentanetetrol. Biochim Biophys Acta. 1987;921:281–291. doi: 10.1016/0005-2760(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 4.Huber R, Wilharm T, Huber D, Trincone A, Burggraf S, Konig H, Rachel R, Rockinger I, Fricke H, Stetter K O. Aquifex pyrophilus gen. nov. sp. nov. represents a novel group of marine hyperthermophilic hydrogen-oxidizing bacteria. Syst Appl Microbiol. 1992;15:340–351. [Google Scholar]
- 5.Ishii M, Kawasumi T, Igarashi Y, Kodama T, Minoda Y. 2-Methylthio-1,4-naphthoquinone, a new quinone from an extremely thermophilic hydrogen bacterium. Agric Biol Chem. 1983;47:167–169. doi: 10.1128/jb.169.6.2380-2384.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ishii M, Kawasumi T, Igarashi Y, Kodama T, Minoda Y. 2-Methylthio-1,4-naphthoquinone, a unique sulfur-containing quinone from a thermophilic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. J Bacteriol. 1987;169:2380–2384. doi: 10.1128/jb.169.6.2380-2384.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawasumi T, Igarashi Y, Kodama T, Minoda Y. Hydrogenobacter thermophilus gen. nov., sp. nov., an extremely thermophilic, aerobic, hydrogen-oxidizing bacterium. Int J Syst Bacteriol. 1984;34:5–10. [Google Scholar]
- 8.Koga Y, Nishihara M, Morii H, Matsushita M A. Ether polar lipids of methanogenic bacteria: structures, comparative aspects, and biosyntheses. Microbiol Rev. 1993;57:164–182. doi: 10.1128/mr.57.1.164-182.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pitulle C, Yang Y, Marchiani M, Moore E R B, Siefert J L, Aragno M, Jurtshuk P, Jr, Fox G E. Phylogenetic position of Hydrogenobacter. Int J Syst Bacteriol. 1994;44:620–626. doi: 10.1099/00207713-44-4-620. [DOI] [PubMed] [Google Scholar]
- 10.Shiba H, Kawasumi T, Igarashi Y, Kodama T, Minoda Y. The CO2 assimilation via the reductive tricarboxylic acid cycle in an obligately autotrophic, aerobic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. Arch Microbiol. 1985;141:198–203. [Google Scholar]
- 11.Sprott G D, Ferrante G, Ekiel I. Tetraether lipids of Methanospirillum hungatei with head groups consisting of phospho-N,N-dimethylaminopentanetetrol, phospho-N,N,N-trimethylaminopentanetetrol, and carbohydrates. Biochim Biophys Acta. 1994;1214:234–242. doi: 10.1016/0005-2760(94)90069-8. [DOI] [PubMed] [Google Scholar]
- 12.Yoon K-S, Ishii M, Igarashi Y, Kodama T. Purification and characterization of 2-oxoglutarate:ferredoxin oxidoreductase from a thermophilic, obligately chemolithoautotrophic bacterium, Hydrogenobacter thermophilus TK-6. J Bacteriol. 1996;178:3365–3368. doi: 10.1128/jb.178.11.3365-3368.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon K-S, Ishii M, Kodama T, Igarashi Y. Purification and characterization of pyruvate:ferredoxin oxidoreductase from Hydrogenobacter thermophilus TK-6. Arch Microbiol. 1997;167:275–279. doi: 10.1007/s002030050443. [DOI] [PubMed] [Google Scholar]
- 14.Yoon K-S, Ishii M, Kodama T, Igarashi Y. Carboxylation reactions of pyruvate:ferredoxin oxidoreductase from Hydrogenobacter thermophilus TK-6. Biosci Biotechnol Biochem. 1997;61:510–513. [Google Scholar]