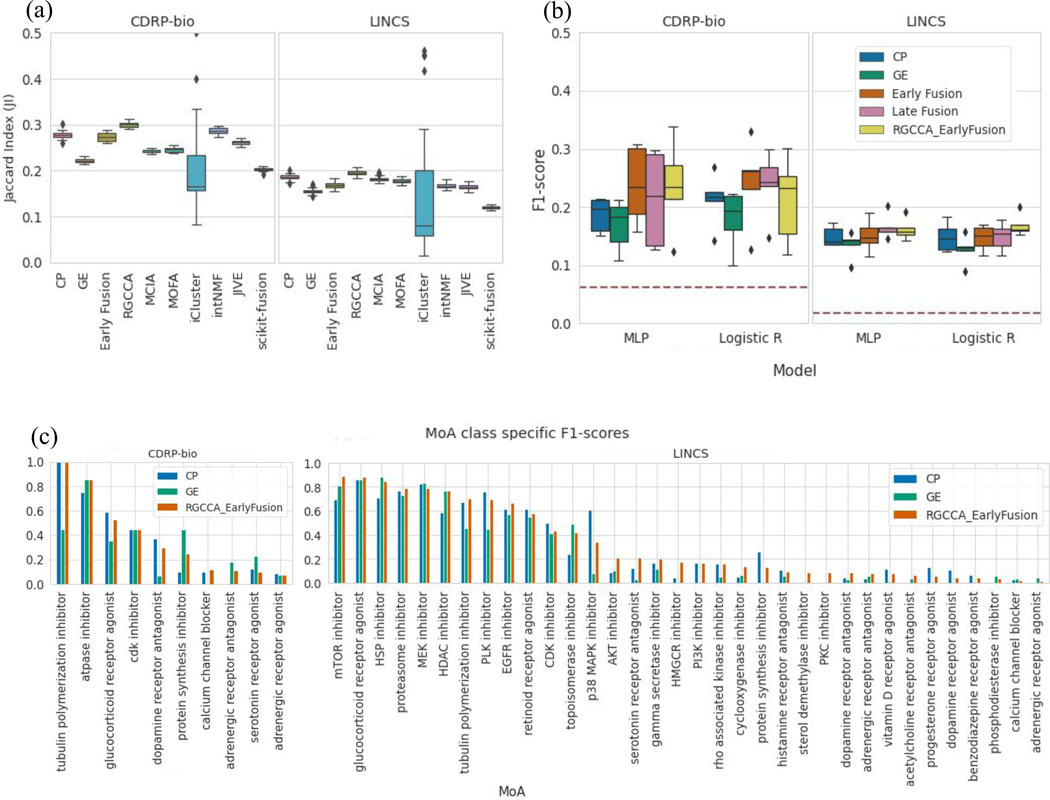
Figure 4. Utilizing Complementary Information: Data Integration for MoA cluster retrieval and class prediction in compound datasets.
An application using the complementary subspaces: integrating multimodal data for mechanism of action (MoA) unsupervised clustering retrieval (a) and supervised prediction (b): (a) Benchmarking of data integration methods on the task of clustering compounds by their MoA categories. Distribution of the Jaccard Indices (one per MOA class; higher is better) computed between the clusters identified by the different integration methods28 and the ground-truth MoA clusters. Regularized Generalized Canonical Correlation Analysis (RGCCA) improves MoA retrieval for both CDRP-bio and LINCS datasets. Distributions are presented as boxplots, with the center line being median, box limits being upper and lower quartiles and whiskers being 1.5× interquartile range; n=16 (CDRP-bio), n=57 (LINCS) (b) MoA classification of the two compound datasets (CDRP-bio and LINCS) using gene expression, morphology and their integration to predict the mechanism of action of compounds. Classification performance (weighted F1-score) for the multilayer perceptron (MLP) and Logistic Regression classifiers using each data modality alone, the two early and late fusion strategies explained in the main text, and the early fusion of modalities after application of RGCCA on the feature space of both modalities. Chance-level predictions for each dataset are shown as a horizontal red line on each dataset plot. Distributions are presented as boxplots, with the center line being median, box limits being upper and lower quartiles and whiskers being 1.5× interquartile range; n=k=5. (c) Class-specific F1-scores are shown based on the MLP model for 16 MoA categories of CDRP-bio (left, where the 4 out of 16 MoA categories that resulted in zero F1-scores after fusion are excluded) and for LINCS (right, where the 23 out of 57 MoA categories that resulted in zero F1-scores after fusion are excluded).