Figure 3.
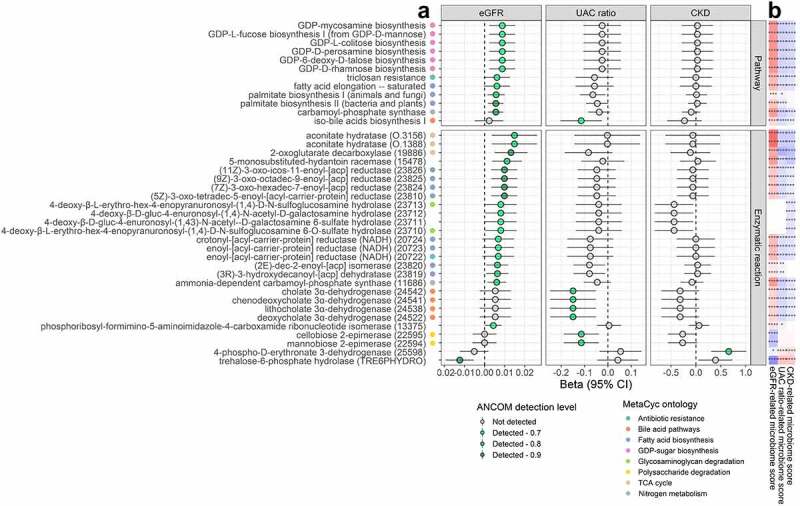
Gut microbiome functions associated with kidney traits in the Hispanic Community Health Study/Study of Latinos (N = 2,438). (a) We used ANCOM2 to identify MetaCyc pathways and enzymatic reactions for which abundance was associated with eGFR, UAC ratio, or CKD. ANCOM2 models were adjusted for age, sex, field center, Hispanic/Latino background, U.S. nativity, antibiotics use, Bristol stool type, income, educational attainment, cigarette smoking, alcohol use, AHEI2010, predicted sodium intake, report of low-sodium diet, predicted protein intake, report of high protein/low carb diet, protein supplement use, total physical activity, BMI, waist-to-hip ratio, systolic blood pressure, diastolic blood pressure, triglycerides, HDL cholesterol, fasting glucose, hypertension medication, diabetes medication, and lipid-lowering medication. For each pathway/reaction associated with at least one kidney trait in ANCOM2 (detection level ≥0.7), we show the estimated effect size (beta) and 95% confidence interval for eGFR (ml/min/1.73 m2), log UAC ratio, and CKD from multivariable linear regression models, with clr-transformed pathway/reaction abundance as outcomes, adjusting for same covariates as used in ANCOM2. Pathways/reaction names are annotated with colored circles according to their ontology in the MetaCyc database (only for categories with ≥2 constituents). (b) Spearman correlations of kidney trait-related microbiome scores and clr-transformed pathway/reaction abundance. Scores were derived by Z-score standardizing the clr-transformed abundance of species associated with a given kidney trait, followed by summing/subtracting species that were positively/negatively related to the trait.
