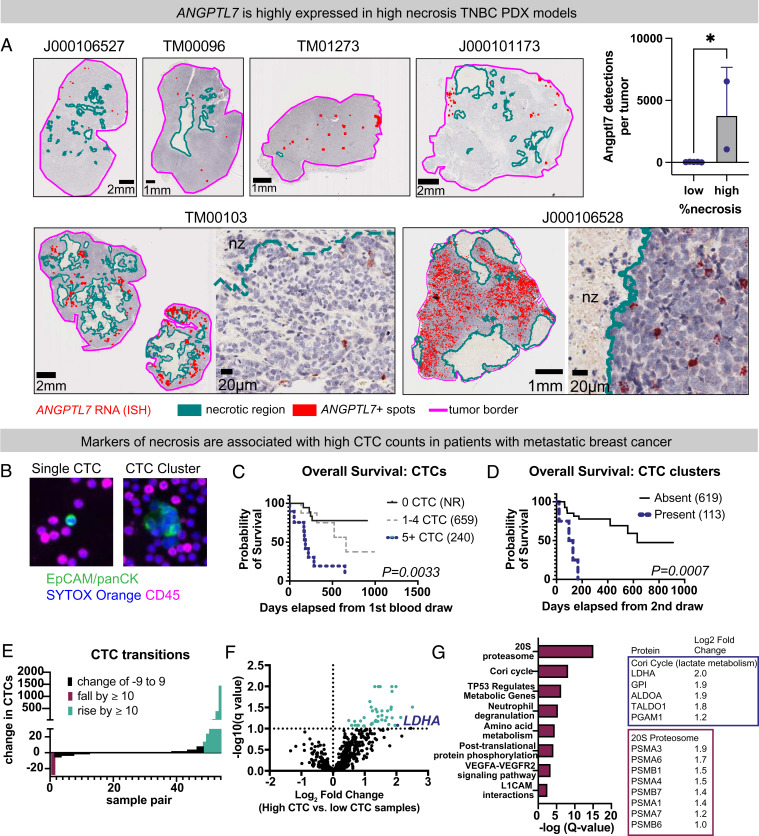
Fig. 6.
ANGPTL7 is expressed in high necrosis human triple-negative breast cancers and necrosis markers correlate with CTC dissemination in breast cancer patients. (A) ANGPTL7 RNA ISH performed on tumor sections from human breast PDX tumors transplanted into NSG mice. All tumor models were invasive ductal carcinomas and confirmed triple-negative for ER, PR, and HER2 on PDX tumor sections. Qupath detections shown. Turquoise: necrotic region, red: ANGPTL7-positive detections, pink: tumor border. (Insets) show raw ANGPTL7 RNA ISH in red. (Right) Bar graph comparing tumors with low necrosis (five sections from four models) and high necrosis (two sections from two models). High necrosis defined as >15% necrosis by area. P-value determined by unpaired t test. (B) Representative single CTC and CTC cluster from clinical vignette in SI Appendix, Fig. S6. EpCAM/pan-CK, epithelial cells; CD45, immune cells; SYTOX Orange, nucleic acid stain. (C) Overall survival according to CTC enumeration at baseline. Median overall survival reported in parentheses. NR = not reached. P-value determined by Mantel–Cox log-rank test. (D) Overall survival according to presence or absence of CTC-clusters. Landmark analysis from 2nd blood draw was performed. Median overall survival reported in parentheses. P-value determined by Mantel–Cox log-rank test. (E) Waterfall plot showing changes in CTC abundance between time points, sorted by magnitude. (F) Tandem-mass tag mass spectrometry of high vs. low-CTC samples. Volcano plot shows the 46 plasma proteins enriched in three high-CTC samples compared with 13 low CTC samples. Proteins with Q-values less than 10%. False discovery rate determined by two-stage set-up method by Benjamini, Krieger, and Yekutieli. (G) Metascape analysis of CTC-associated proteins reported by -log Q-value. (Right) Proteins with fold-enrichment that make up core enrichments for Cori cycle and 20S proteasome. *P <0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.