FIGURE 2.
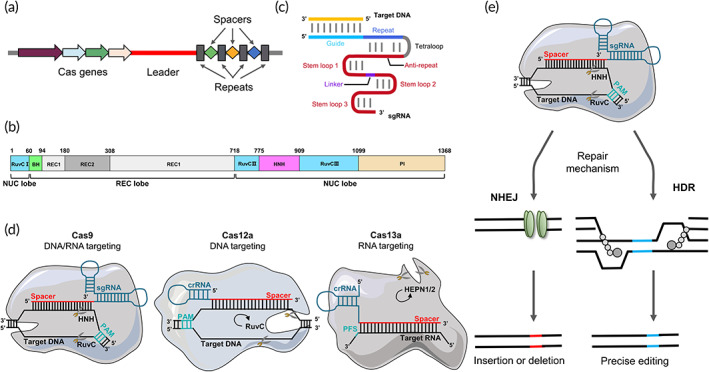
The structure of the Class II CRISPR–Cas systems and gene editing mechanism (with CRISPR–Cas9 as an example). (a) Typical structure of CRISPR locus. The CRISPR gene sequence is mainly composed of the leader, repeats and spacers. The leader sequence is located upstream of the CRISPR gene and is considered as the promoter of the CRISPR sequence. The repeats are about 20–50 bp base length and the transcription products can form hairpin structures. The spacers are exogenous DNA sequence that are captured by the bacteria. (b) The domain organization of SpCas9 consists of NUC lobe and REC lobe. BH, Bridge helix. (c) Schematic representation of the sgRNA:target DNA complex. Artificially designed target sequences of sgRNAs function as crRNA‐tracRNA complexes, which can direct Cas9 proteins to specifically cleave target genes. (d) Schematic representation of representative Cas proteins from different families (shown are Cas9, Cas12a, and Cas13a). In CRISPR–Cas9, the sgRNA‐encoded spacer binds to the target dsDNA near the PAM. Base pairing activates the HNH and RuvC nuclease structural domains, which separate the two strands. In CRISPR–Cas12a, the crRNA‐encoded spacer binds to the target base and activates the RuvC nuclease, cleaving both strands with multiple‐turnover general ssDNase activity (arrow). In CRISPR–Cas13a, the target sequence is RNA. Correct base‐pairing activates HEPN nuclease general ssRNase activity (arrow). (e) Genome editing using CRISPR–Cas9. The Cas9 nuclease binds to the sgRNA, which in turn is directed to the target DNA by complementary base pairing. PAM sequence (NGG, NAG) must be present in the anterior segment of the target sequence. Cleavage of the next double‐stranded DNA (dsDNA) triggers the error‐prone nonhomologous end joining (NHEJ) or homologous directed repair (HDR) mechanism
