Fig. 6.
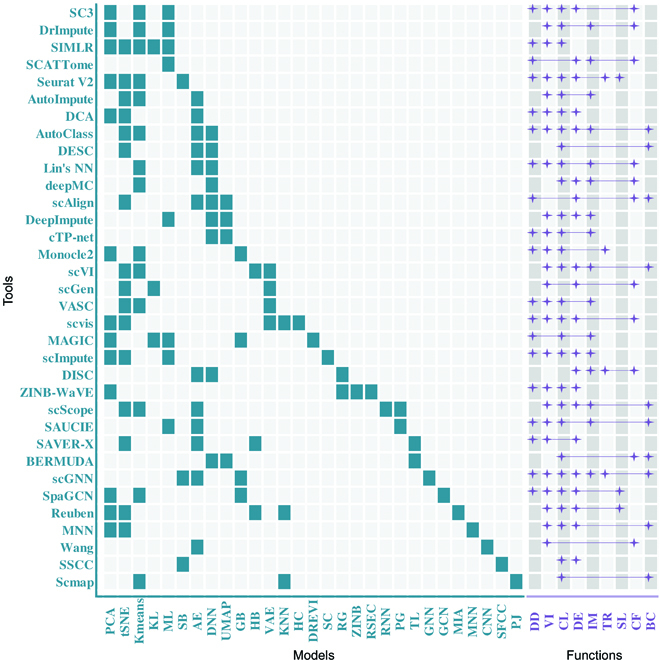
ML and DL techniques used in single-cell sequencing data analysis. Analysis tools are represented on the vertical axis, models are shown on the horizontal axis in green, and functions are implemented by tools in purple. KL, kernel learning; ML, metric learning; SB, statistic-based methods; AE, autoencoder; DNN, deep neural network; GB, graph-based methods; HB, hierarchical Bayesian; VAE, variational autoencoder; KNN, k-nearest neighbor; HC, hierarchical clustering; DREVI, density reweighted visualization; SC, spectral clustering; RG, regression; ZINB, zero-inflated negative binomial; RSEC, resampling-based sequential ensemble clustering; RNN, recurrent neural network; PG, PhenoGraph, a graph-based method; TL, transfer learning; GNN, graph neural network; GCN, graph convolutional network; MIA, multimodal intersection analysis; MNN, mutual nearest neighbors; CNN, convolutional neural network; SFCC, subsampling–featuring–clustering–classification; PJ, projection algorithm; DD, data denoising; VI, visualization; CL, clustering; DE, differential expression genes analysis; IM, imputation; TR, trajectory analysis; SL, spatial location; CF, classification; BC, batch correction.
