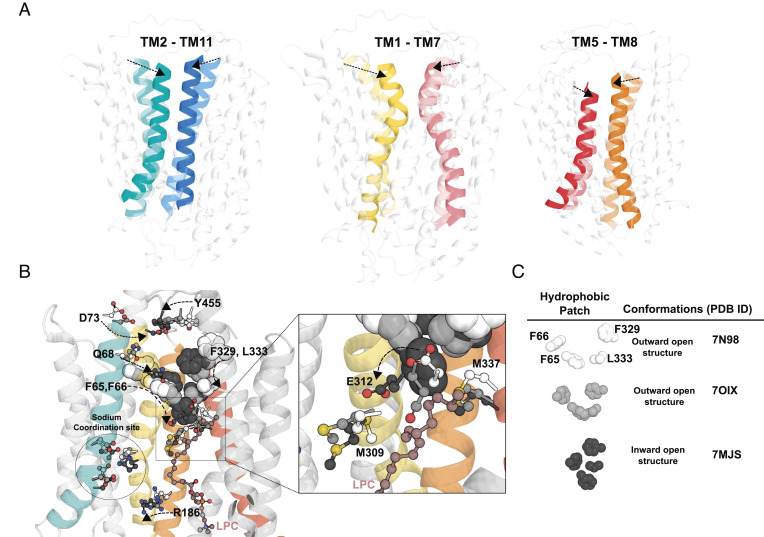
Fig. 6.
Conformational changes during LPC flipping. (A) Movement of the TM2-TM11, TM1-TM7, and TM5-TM8 from the superposition of the three determined cryo-EM structures, outward-open (Left panel, Protein Data Bank (PDB): 798N), outward-open occluded (Middle panel, PDB: 7OIX), and inward-open (Right panel, PDB: 7MJS) conformations. Left panel showing TM2 and TM11, Middle panel showing TM1 and TM7, Right panel showing TM5 and TM8, with dotted arrows showing conformational changes in the indicated TMs from the outward-open structure to inward-open structure. (B) Leftmost structure showing residues of TM1 (D73, Q68, F65, and F66) and TM7 (F329, L333, and E312) are seen to move toward each other from the outward-open structure 7N98 to the inward-open structure 7MJS (see dotted line arrows). As the residues from both TM1 and TM7 move toward each other to form the inward-open structure, E312 is seen to move from M337 to M309 (see enlarged area indicated by a square). (C) Changes in the relative positions of the four hydrophobic residues F65, F66, F329, and L333 from the outward-open structure 7N98 to the occluded structure 7OIX resulting in the formation of a hydrophobic plug in the inward-open structure 7MJS. Residues D73, Q68, Y455, E312, M309, and M337 are represented in sticks and colored white for outward-open structure 7N98, light gray for outward-open occluded structure 7OIX and dark grey for inward-open structure 7MJS (see Figure Legend). An LPC molecule that is found in the inward-open structure, 7MJS, is represented in maroon ball and sticks.