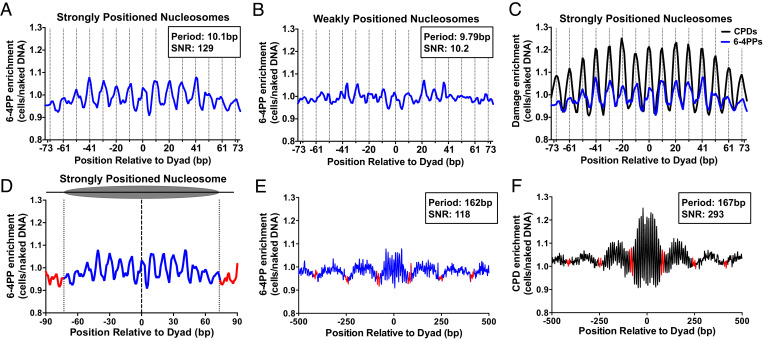
Fig. 2.
Rotational positioning of DNA in nucleosomes regulates 6-4PP formation in yeast and human cells. (A and B) Rotational periodicity of 6-4PPs within ~10,000 strongly positioned (A) and ~7,500 weakly positioned (B) yeast nucleosomes, derived from a published map of yeast nucleosome positions (29). Dashed lines correspond to outward rotational settings. UVDE-seq reads for 6-4PPs in UV-irradiated yeast cells (UV 0 h) were normalized to UVDE-seq reads for 6-4PPs from naked DNA controls, in order to account for DNA sequence biases in nucleosomal DNA and calculate 6-4PP enrichment. The peak periodicity and signal-to-noise ratio (SNR), which quantifies the strength of the periodicity (11, 30), are shown for 6-4PP enrichment in nucleosomes. (C) 6-4PPs data from panel A plotted against CPD enrichment within strongly positioned yeast nucleosomes, using published CPD-seq data (10). (D) Analysis of translational periodicity of 6-4PP enrichment in nucleosomal DNA (blue) relative to linker DNA (red) in strongly positioned (29). Dashed lines at −73 and +73 represent the border between nucleosomal and linker DNA. (E and F) Translational periodicity of E 6-4PP or F CPD enrichment in a 1,000-bp window centered on strongly positioned nucleosomes (29). Red indicates likely location of linker DNA. Peak translational periodicity and its SNR was calculated using mutperiod (30).