Abstract
Structural variations (SVs) are a key type of cancer genomic alterations, contributing to oncogenesis and progression of many cancers, including colorectal cancer (CRC). However, SVs in CRC remain difficult to be reliably detected due to limited SV-detection capacity of the commonly used short-read sequencing. This study investigated the somatic SVs in 21 pairs of CRC samples by Nanopore whole-genome long-read sequencing. 5200 novel somatic SVs from 21 CRC patients (494 SVs / patient) were identified. A 4.9-Mbp long inversion that silences APC expression (confirmed by RNA-seq) and an 11.2-kbp inversion that structurally alters CFTR were identified. Two novel gene fusions that might functionally impact the oncogene RNF38 and the tumor-suppressor SMAD3 were detected. RNF38 fusion possesses metastasis-promoting ability confirmed by in vitro migration and invasion assay, and in vivo metastasis experiments. This work highlighted the various applications of long-read sequencing in cancer genome analysis, and shed new light on how somatic SVs structurally alter critical genes in CRC. The investigation on somatic SVs via nanopore sequencing revealed the potential of this genomic approach in facilitating precise diagnosis and personalized treatment of CRC.
Author summary
Structural variants contribute to oncogenesis and progression of colorectal cancer, but they remain difficult to be reliably detected. Aiming at obtaining a comprehensive picture of somatic SVs in CRC, we perform long-read nanopore sequencing on CRC tumor samples and their matched para-carcinoma tissues. Our results show long-read sequencing precisely and reliably detects 494 somatic SVs per sample, which are significantly more than previously reported short-read sequencing based studies. We find large scale inversions (>10 kbp) that are frequently difficult to be detected by short-read sequencing and alter the expression or structure of key tumor suppressor genes (including APC and CFTR). A novel gene fusion RNF38-RAD51B is also identified, and we find it functionally acts to enhance migration, invasion, and metastasis capabilities of colorectal cancer cells. Although the molecular mechanisms and clinical relevance of the inversions and gene fusions need to be further studied, our work presents a relatively complete SV landscape of CRC, and providing a genetic basis for CRC’s personalized medicine.
Introduction
Colorectal cancer (CRC) is the third most common malignancy with over 1.8 million new cases and 0.86 million deaths worldwide in 2018 [1]. The development and progression of CRC are largely attributed to genetic alterations, such as structural variations (SVs), single nucleotide variations (SNVs), and epigenetic changes. Among these genetic alterations, the SVs that affect gene expression and function via gene amplification or deletion, gene structure disruption, and gene fusion, are prevalent in CRC [2,3], and have been examined in several studies by copy number variation (CNV) arrays and short-read sequencing [4–7]. These studies identified copy number alterations of oncogenes (including KRAS and MYC), deletions of tumor suppressors (such as FHIT, PTEN, SMAD2 and SMAD4), and recurrent R-spondin fusions [8,9]. However, CNV arrays are incapable of determining precise positions of most of SVs, and short-reads sequencing is inefficient in detecting long, complex, or repetitive-region located SVs [10–12]. Thus, precise and detailed detection of SVs in CRC still remains as a challenge [13,14].
Long-read sequencing technologies can generate long continuous reads (length over tens of kilobase pairs (kbp)), possess increased reliability and sensitivity in SVs detection [15]. Pacific Bioscience (as called single-molecule read-time (SMRT) sequencing or PacBio sequencing) and Oxford Nanopore Technologies (ONT, or nanopore sequencing) are the two major strategies of long-read sequencing [16]. Unlike short-read sequencing, PacBio and nanopore sequencing generate reads directly from native DNA (without ultrasonic / enzymatic fragmentation and PCR amplification), avoid the difficulty of detecting variants in genome regions with repeat content or atypical GC content [17]. The advantages of long-read sequencing in studying human diseases were highlighted in several studies. For instance, a pentanucleotide repeat expansion in SAMD12 that may cause familial cortical myoclonic tremor with epilepsy was identified using nanopore sequencing [18]. This type of repetitive-region residing SVs were difficult to be analyzed by short-read sequencers [18]. In addition to this, long read sequencing identified the leukoencephalopathy-related GGC repeat expansions, X-linked Dystonia-Parkinsonism-related SINE-VNTR-Alu retrotransposon insertions [19], and these variants were previously missed by short read sequencing. Moreover, long read sequencing realized fast and low-cost genome sequencing of pathogen, such as SARS-CoV-2 [20].
In addition to hereditary disease, long-read sequencing also facilitates the studies of cancer genome. Using nanopore sequencing, a complex KLHDC2-SNTB1 fusion (larger than 10 kbp) composed by three separate chromosome regions was discovered in a breast cancer cell line (SK-BR-3) using nanopore sequencing [11]. In lung adenocarcinoma, a novel class of complex SVs consisting of several small/ middle-sized SVs, were identified via the latest nanopore PromethION sequencer [21]. Given the advantages of long read sequencing, novel large-scale and / or complex SVs that affecting the structure and expression of key oncogenes or tumor suppressor genes, repetitive-region residing SVs that may causing genomic instability (such as transposable element) or contributing to tumor progression, and tumor-promoting gene fusions may be efficiently detected, which would provide a more comprehensive understanding of the genomic aberrations of CRC and further in-depth study of their biology functions.
Here, using long-read whole genome sequencing to analyze CRC tumors from 21 patients, we (1) precisely and reliably detected somatic SVs across the cancer genomes, (2) showed the representative large-scale inversions that altered the expression or structure of key tumor suppressor genes, such as APC and CFTR, in CRC; (3) discovered a novel gene fusion RNF38-RAD51B that could increase the migration, invasion and metastasis ability of CRC cells.
Material and methods
Ethics statement
This study was conducted according to the Helsinki human subject doctrine and was approved by the Huazhong University of Science and Technology review board and Ethics Committee (IORG No. IORG0003571, 2020-S197), written consents to participate was acquired from all the patients.
Sample collection and Oxford nanopore sequencing
21 pairs of tumor samples and matched para-carcinoma samples were obtained from the surgically removed tumor tissues and adjacent intestinal tissues (>6 cm from tumor tissues) of CRC patients in Wuhan Union Hospital, and stored at -80°C. All the samples were analyzed and sequenced using long-read Nanopore sequencing, short-read whole exome sequencing and RNA sequencing. Genomic DNA from each sample was extracted by sodium dodecyl sulphate method. DNA was shared to > 20kb by Covaris g-tude. Then genomic DNA libraries were constructed according to the manufacturer’s instructions by using the Ligation Sequencing kit 1D (SQK-LSK109). The prepared libraries were loaded into R9.4(1D) flow cells and then sequenced on the PromethION sequencer (ONT, UK). Then Guppy (version: 2.0.8) was used to perform basecalling on fast5 files to generate FASTQ format files.
Alignment and SVs calling
All the reads from ONT sequencing were aligned to the human reference genome with only major chromosomes 1–22 and X, Y from NCBI (ftp://ftp-trace.ncbi.nih.gov/1000 genomes/ftp/technical/reference/human_g1k_v37.fasta.gz) using NGMLR (v0.2.7) with default parameters. Samtools (v1.9) was used to compute alignment ratio and mapping identity by analyzing bam files. Structural Variations were called using Sniffles v1.0.8 with minimum reads supporting number 2 and minimum SV size 50bp. In order to obtain high-quality SVs in tumor and normal samples, only SVs supported by at least 0.3 folds of average sequencing depth were retained.
Somatic SVs (present in tumor but not in normal samples SV calls) were obtained by comparing high quality tumor samples SVs passed above filtering thresholds with normal samples SVs only supported by two or more reads. This strategy is to improve the recall rate of normal samples SVs to improve the reliability of somatic SVs. Tumor and matched normal sample SVs were merged using svmerge (https://github.com/GrandOmics/svmerge) with a maximum distance of 1000bp for all types SVs, 40% reciprocal overlaps for deletions, inversions and duplication and difference in SV length less than 20%. We used svhawkeyes (https://github.com/yywan0913/SVhawkeye) for the manual curation of unfiltered somatic SVs. The reads alignment images of each unfiltered somatic SVs were generated by svhawkeyes from alignment files and manually checked. Somatic SVs that appear in both cancer and paired normal samples were identified as false positive. Finally, all the somatic SVs were merged into an integrated call set. SVs with upstream and downstream genes were annotated in the segdup (UCSC golden path hg19), rmsk (UCSC golden path hg19), dgv (2016-05-15), 1000 Genome Project (phase 3), gnomAD (2.1.1), and COSMIC databases (v70) using annovar (2017-07-17). Insertions were further annotated as tandem repeats or known repeat classes using TRF (4.09) and RepeatMasker (4.1).
Whole exome sequencing and variants calling
Sheared genome DNA from each tumor and normal sample was used for library preparation. Exome DNA was captured using the XGen Exome Research Panel v1.0 51Mb kit and sequenced using the Illumina NovaSeq platform with 150 bp paired-end sequencing mode. The sequencing depth of each sample was above 200X. Bam files were generated using sentieon DNA pipelines (sentieon-genomics-201808.01) including alignment, removing duplications, sorting and local realignment following the Broad Institute’s best practices. Somatic mutations and Indels were detected by using Sentieon TNscope from co-realigned tumor and normal BAM files with dbSNP 138 in target intervals. All somatic mutations and Indels were annotated in the dbSNP 147, clinvar (2017-05-01), ExAC (2016-04-23), 1000 Genome Project (phase 3), gnomAD (2.1.1), InterVar (2017-02-02) and COSMIC databases (v70) using ANNOVAR (2017-07-17).
Transcriptome sequencing and quantification of gene expression level
Sequencing libraries were generated using NEBNext Ultra RNA Library Prep Kit for Illumina (NEB, USA) according to the manufacturer’s instructions. AMPure XP system (Beckman Coulter, Beverly, USA) was used to purify the library fragments and 3 μl USER Enzyme (NEB, USA) was used for size selection (250~300 bp). The library preparations were sequenced on an Illumina Hiseq platform with 150 bp paired-end model, and at least 6 G of clean data were generated for each sample. Paired-end reads were aligned to the reference genome using Hisat2 (v2.0.5). Reads counts were calculated using FeatureCounts (v1.5.0-p3). Differential expression analysis was performed using the edgeR R package (v 3.18.1) and significance was defined as adjusted P-value < 0.05 and foldchange > 2.
Novel gene fusions identification
Fusion gene usually caused by reasons such as chromosome translocation, inversion and deletion. Two genes containing two breakpoints of the same SV respectively were selected as candidate fusion gene. star-fusion(1.2.0) was used to detect fusion genes from Illumina RNA sequencing data with—annotate;—examine_coding_effect;—FusionInspector inspect;—denovo_reconstruct;—min_junction_reads 1;—min_sum_frags 2. Fusion genes predicted by structural variation and expressed in the RNAseq data were further used for manual curation from both nanopore whole genome sequencing and Illumina RNA sequencing alignments. Primers were designed to span fusion junction and were validated by PCR and Sanger sequencing.
Primer sequences
| Breakpoint 1 | Breakpoint 2 | |||
| Forward (5’ to 3’) | Reverse (5’ to 3’) | Forward (5’ to 3’) | Reverse (5’ to 3’) | |
| APC inversion | TGGGTATCAGATCTCTATAGGCTGT | GCACTTCTATGTATGTGTCAGGG | ACCAGAAGGCAGGGTCATTG | CCCAGCAAGCAAGGAAGTTG |
| CFTR inversion | ACAAATTCCAAGACTTACTGGCA | TGGTCACTGGCTTGTTGAGA | GACATGATCCTTTTGCAGCCT | TGTGCCCACAGTTCAAACCT |
| RNF38-RAD51B gene fusion | TGGTTTGGCTACTTTCCCTCT | GCAGGGGTACTCAAAGTCCC | ||
| SMAD3-SHISA6 gene fusion | GAAGCCAAAACACCGGACAC | TCATACTTCTGGGGCTGGGA | TGGCTGAAGGTCTGTTTTGT | ACAGAGAAGCCAAGAAGCCA |
Cell lines
HEK293T (human embryo kidney cell line), LoVo (human colon adenocarcinoma cell line) and HCT116 (human rectum adenocarcinoma cell line) cells were purchased from the American Type Culture Collection (Rockville, MD, USA) and maintained in Dulbecco’s Modified Eagle medium (Hyclone, Logan, UT, USA), supplemented with 10% fetal bovine serum (Sciencell, Carlsbad, CA, USA) at 37°C under 5% CO2 in a cell incubator.
Establishment of RNF38-RAD51B overexpressing cell lines
The cDNA of RNF38 and RAD51b was amplified from a cDNA library of HCT116 cells, then cloned into pLenti-puro lentiviral reporter plasmid to form a RNF38-RAD51b overexpression vector. The overexpression vector was confirmed by PCR and Sanger sequencing (sequences of the primer pair are listed below). Then, the lentivirus vector was obtained by co-transfecting HEK293T cells with pLenti-puro-RNF38-RAD51b, psPAX2 packaging, and pMD2.G enveloped plasmids according to the manufacturer’s instructions. HCT116 and LoVo cells were infected by filtered lentivirus (pLenti-puro-vector or pLenti-puro-RNF38-RAD51b) with polybrene (8 μg/mL) and then selected by puromycin (1 μg/mL) for 1 week. The expression level of RNF38-RAD51B fusion gene was measured using western-blot.
Transwell migration and invasion assays
The migration and invasion assays of RNF38-RAD51B overexpressing HCT116 and LoVo cells were assessed using 8.0-μm pore size transwell inserts. For migration assay, cells were seeded to inserts and cultured for 15 (LoVo cells) or 30 (HCT116 cells) hours. For migration assay, cells were seeded to Matrigel-coated inserts (invasion assay) and cultured for 36 (LoVo cells) or 48 (HCT116 cells) hours Then, the cells on the underside of the inserts were fixed and stained with crystal violet, and counted by microscope. Each experiment was repeated for thrice.
Animal studies
Six weeks old male BALB/c nude mice purchased from Beijing HFK Bioscience Co., Ltd were used for the animal studies. 1 × 106 RNF38-RAD51B overexpressing HCT116 cells were injected into the livers of the nude mice via the splenic vein (eight mice per group). After six weeks, the mice were euthanized by excessive anesthesia and the livers were collected. Then, the liver tissues were sectioned, stained with hematoxylin and eosin (H&E), and assessed by quantifying the number of metastatic lesions by a microscope.
Results
Nanopore sequencing of CRC samples
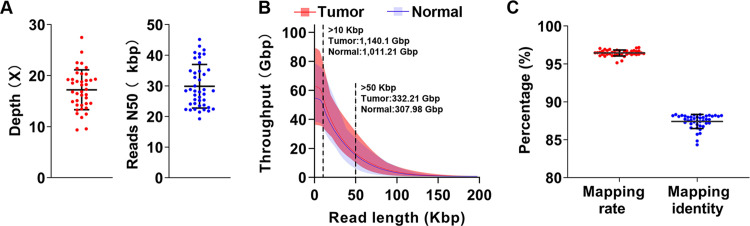
We generated whole-genome long-read sequence data from 21 CRC patients (S1 Table) using PromethION (Oxford Nanopore Technologies) nanopore sequencers. All the patients were at stage II (n = 13) or stage III (n = 8), and five of them were of high-level microsatellite instability (MSI-H). All samples were also analyzed by short-read whole exome sequencing (WES) and RNA-seq to obtain SNVs and gene transcriptional data, respectively. We obtained over 51 billion bases (>17X in depth) long-read data per sample with a mean read N50 of 30,211 bp (range from 19,238 bp to 45,166 bp; 94% of reads were ≥10 kbp) (Fig 1A and 1B and S2 and S3 Tables). The maximum read length and the N50 length of the obtained reads were 897,996 bp and 42,969 bp, respectively, consistent with previously reported PromethION data [21], but longer than those generated by MinION platform [11,15]. With NGMLR [22], 96.3% of the reads were mapped to the reference genome (human G1Kv37) with the mean mapping intensities of 87.4% (Fig 1C).
Fig 1. Summary of the long-read sequencing data.
(A) The sequencing depth (left) and reads N50 (right) of long-read data obtained from 21 pairs of tumor/ normal samples via nanopore sequencer. (B) Cumulative distribution of total bases (Y axis) over read length (X axis) for tumor/ normal samples. The quantities of bases in reads 10 Kbp+ and 50 Kbp+ were labeled. (C) The mapping rate and identity of long-reads sequencing.
Long-read sequencing identified widespread somatic SVs in colorectal cancer
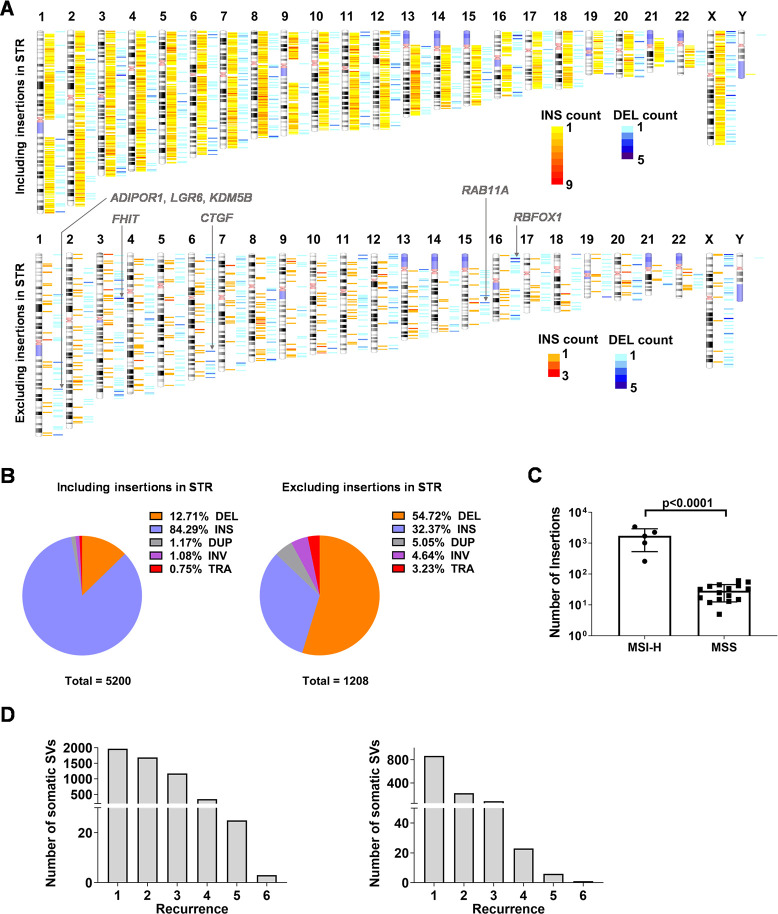
We employed Sniffles [22] for SV calling and identified 817,857 SVs in all the samples (19466 SVs per sample) (S1 Fig), largely consistent with previous studies [22,23]. These SVs were used to map somatic SVs, yielding 14508 unfiltered somatic SVs. After manual curation (see Method), we obtained 494 somatic SVs per tumor sample (in total 5,200 nonredundant somatic SVs), significantly more than previous short-read data in CRC [24,25], likely due to the increased sensitivity of long-read sequencing in detecting SVs [11,26]. The lengths of 98% of SVs were less than 10,000 bp, and the mutual distance of most SVs (~80%) was between 105 ~ 107 bp (S2 Fig). The components of these somatic SVs were 661 (12.7%) deletions, 4,383 (84.3%) insertions, 61 (1.2%) duplications, 56 (1.1%) inversions, and 39 (0.8%) translocations (Figs 2A, upper, 2B, left; and S3). The classification of sequences and loci of these insertions and deletions revealed that most of insertions (95%) occur in MSI-H samples due to the abnormal expansion of short tandem repeat (STR) regions (Figs 2C and S4). After exclusion of insertions in STR regions, 54.72%, 32.37%, 5.05%, and 3.32% of somatic SVs were deletions, insertions, duplications, and translocations, respectively (Figs 2A, lower, 2B, right; and S3). The number of inversions in MSI-H samples was significantly lower than that in MSS samples, and the numbers of other types of SVs were similar across different MSI status and different stages (S5 Fig). Some loci with high frequency were associated with the genes involved in oncogenesis and development of CRC, including alternative splicing factor RBFOX1, tumor suppressor gene FHIT, and several oncogenes such as LGR6, CTGF and RAB11A (Fig 2A). Meanwhile, 62.1% of somatic SVs were detected in at least two samples (Fig 2D). Recurrent insertions were mainly located in STR regions (S6 Fig). In addition, duplications, inversions, and translocations were less likely to be recurrent events, as over 90% of these SVs were singletons (S7 Fig).
Fig 2. Detection of somatic SVs in CRC by long-read sequencing.
(A) Chromosome ideogram showing somatic deletions (DEL) and insertions (INS) identified by long-read sequencing in 21 pairs of CRC samples. (B) Pie chart showing the percentages of different classes of SVs identified from long-read sequencing data including or excluding insertions in STR regions. (C) Quantification of somatic insertions in MSI-H or MSS samples (p<0.0001, Student’s t-test). (D) The quantities of somatic SVs that were detected in multiple samples, including (left) or excluding insertions in STR regions (right). The “Recurrence” on X-axis refers to the number of samples in which an SV is detected.
Characterizations of somatic SVs reveal expanded LINE and SINE insertions in CRC
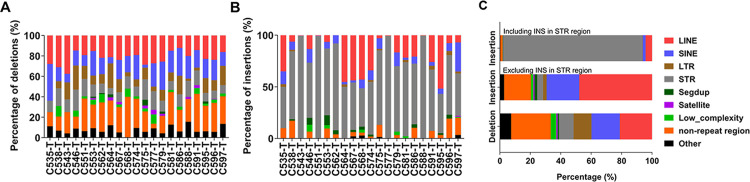
We classified the identified SVs by repeat contents of the variant sequence using RepeatMasker (http://www.repeatmasker.org) to explore the genomic context of somatic SVs. Approximately, half of the deletions were located in tandem-repeats regions or mobile elements (for instance, LINE, SINE and Long terminal repeat (LTR)) (Fig 3A). After exclusion of STR regions, approximately 70% of insertions were mobile elements’ insertions (Figs 3B and 3C and S8), half of which were LINE insertions, indicating aberrant activation of LINE-1 retrotransposons in CRC, consistent with previous reports [27,28].
Fig 3. Sequence analysis of somatic deletions/ insertions.
(A and B) The genomic context of somatic deletions (A) and insertions (B) in each sample obtained from sequence analysis using RepeatMasker. Segdup, segment duplications; Satellite, satellite repeats; Low_complexity, low complexity repeats. (C) The components and proportions of deletions and insertions including or excluding insertions in STR regions.
Large-scale inversions cause dysfunction of tumor suppressors
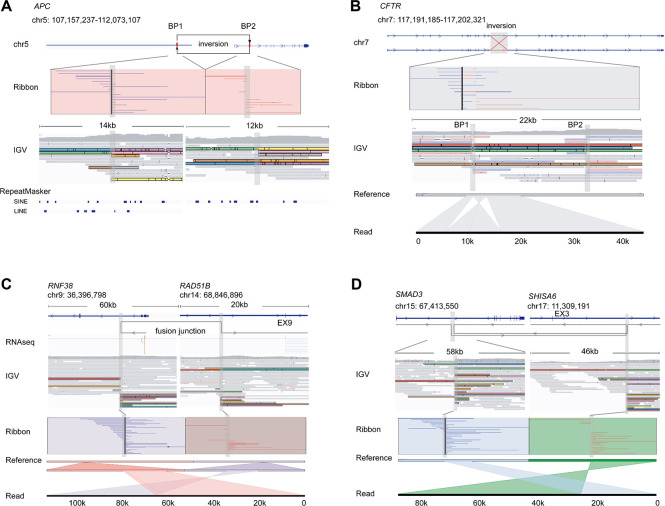
In addition to small SVs, large-scale (> 10 kbp) somatic SVs that affected tumor suppressors through disrupting gene structure to silence them, were also detected by nanopore sequencing. In the sample C546-T, a high-confidence 4.9 Mbp inversion that spanned from chr5: 107,157,237 to chr5: 112,073,107 covering the exon 1 of APC was identified (Fig 4A). We analyzed the PCR products amplified against the sequences spanning across each breakpoint using Sanger sequencing to detail the structure of both breakpoints (S9A Fig). An 8-bp deletion at breakpoint 1 (BP1) was revealed, which resulted in microhomology and might consequently cause the formation of inversion via microhomology-mediated end joining (S10A Fig). RNA-seq results showed that APC expression was sharply decreased (FPKM: 0.296) at mRNA level compared to the paired normal sample (C546-N, FPKM: 2.262). No variant in APC was reported using short-read based WES, which was possibly because the base sequence of inverted exon 1 of APC was unchanged.
Fig 4. Large-scale inversions and gene fusions detected by nanopore sequencing.
(A-D) Reads and structures of the 4,915 kbp inversion spanning exon 1 of APC (A), the 11.2 kbp inversion in CFTR (B), the RNF38-RAD51B gene fusion (C) and the SMAD3-SHISA6 gene fusion (D). For reads alignment, the reads spanning breakpoints were highlighted in colors. For each inversion, the top panel indicates the loci which inversion occurred, and read alignments coving the breakpoints (visualized by Ribbon). The forward and reverse split reads were marked by blue and red, respectively. The middle panel shows reads alignments around breakpoints (visualized by Integrative Genomics Viewer). The bottom panel shows the detailed structure of the SV (visualized by Ribbon). For each gene fusion, the top, middle and bottom panels show translocation loci, reads alignment and split reads, respectively.
Additionally, we identified an 11.2-kbp somatic inversion that spanned from chr7: 117,191,185 to chr7: 117,202,321 involving the exon 11 of CFTR in the sample C564-T (Fig 4B). Notably, four long reads spanned both breakpoints of the inversion (Fig 4B), and covered the complete structure of such a relatively-long inversion. Sanger sequencing of both breakpoints revealed small insertions, deletions, and duplications in the vicinity of both breakpoints, suggesting that this inversion may be generated by microhomology-mediated break-induced replication (S9B and S10B Figs).
Novel gene fusions identified by long-read sequencing
Long read sequencing have proven immensely helpful in detecting gene fusions [29]. For instance, we identified two new rearrangements that possibly resulted in gene fusions, RNF38-RAD51B and SMAD3-SHISA6. For RNF38-RAD51B, the upstream of the intron 3 of RNF38 was connected to the downstream of the intron 8 of RAD51B (Fig 4C). This gene fusion was also detected by RNAseq and confirmed by PCR products encompassing breakpoint junctions (S9C and S11A Figs). The formation of this fusion might change the function of RNF38, which reportedly promotes cancer cell migration and invasion, inhibition of cancer cell apoptosis, and epithelial-mesenchymal transition [30–32]. For SMAD3-SHISA6 (Fig 4D), PCR validated that the downstream of the intron 7 of SMAD3 was connected to the upstream of the intron 3 of SHISA6, while the upstream of the intron 7 of SMAD3 was reversely connected to the downstream of intron 7 of SHISA6 (S9D and S11B Figs). However, this gene fusion was not detected by RNAseq, possibly because of its low expression. Given that SMAD3, a major transcription factor in TGF-β pathway, acts as a tumor suppressor and its functional disruption was positively associated with CRC progression and metastasis [33], this fusion might lead to SMAD3 dysfunction, consequently suppressing the function of TGF-β pathway.
RNF38-RAD51B promotes CRC cell migration, invasion, and metastasis.
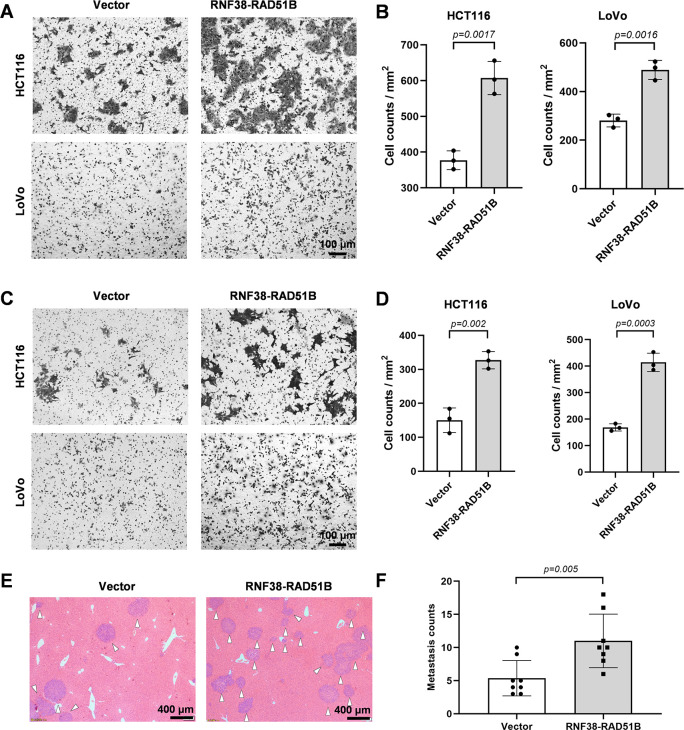
To investigate the oncogenic effects of the RNF38-RAD51B fusion, we cloned the fusion gene and established RNF38-RAD51B overexpressing LoVo (human colon adenocarcinoma cell line) and HCT116 (human rectum adenocarcinoma cell line) cells (S12 Fig). The overexpression of RNF38-RAD51B significantly promoted cell migration and invasion in vitro in transwell assays (Fig 5A–5D). We next examined the in vivo oncogenic roles of the RNF38-RAD51B fusion by intravenously injecting RNF38-RAD51B overexpressing HCT116 cells into nude mice. The metastasis of tumor cells into the livers was observed (Fig 5E and 5F); the number of metastatic loci were two-time higher than that in the control (injected with empty-vector expressing cells). These results demonstrate that RNF38-RAD51B fusion enhances CRC cells’ ability of migration, invasion and metastasis.
Fig 5. RNF38-RAD51B promotes cell migration, invasion and CRC metastasis.
(A, B) Representative images (A) and statistical results (B) of transwell migration assay of RNF38-RAD51B overexpressing CRC cells (three repetitions per group). (C, D) Representative images (C) and statistical results (D) of transwell invasion assay of RNF38-RAD51B overexpressing CRC cells (three repetitions per group). (E, F) Representative H&E-staining images (E) and counts (F) of metastatic tumors in the liver of the xenograft mice intravenously injected with RNF38-RAD51B overexpressing HCT116 cells (eight mice per group). p < 0.05 are statistically significant (students’ t-test).
Discussion
Structural variations are deemed as oncogenic organizers that alter expression and function of oncogenes or tumor suppressors [5]. However, due to the short read length caused ambiguous alignment, commonly used short-read sequencing strategies are ineffective in breakpoints phasing, and complex or long SVs detection and reconstruction [34] Yet, large amount of hidden structural variations in human genomes need to be further identified [15,35]. In this study, we applied nanopore long-read sequencing in 21 pairs of CRC samples, detected approximately twice numbers of somatic SVs in each sample than using short-read sequencing [24,25], and many of them were related to known oncogenes and tumor suppressors. We further investigated the types and components of SVs in CRC, and identified multiple SV hotspots that were associated with CRC-associated genes. This is the first study that employed long-read sequencing to investigate SVs in human CRC samples.
The majority of clinically-used precision therapeutics approaches for colorectal cancer, such as food and drug administration (FDA) approved MSK-IMPACT (Memorial Sloan Kettering Cancer Center) and FoundationOne CDx (Foundation Medicine, Inc) tests, used short-read capture sequencing or amplicon sequencing to detect cancer-relevant and/ or drug-targetable mutations as treatment indicators [36,37]. However, patients might not benefit from short-read capture sequencing or amplicon sequencing if their treatment indicators are SVs [38] since SVs (especially large-scale SVs) may span over one or more exons without any change in their sequence, it is highly possible that these exon-spanning SVs would be missed if using capture sequencing or amplicon sequencing. For instance, the inversions in APC and CFTR clearly altered the structure (including coding regions) of both genes, but were not detected by WES. Thus, detection of such SVs would be valuable to cancer precision therapeutics. Compared to short-read capture sequencing, long reads sequencing are advantageous in capturing large, complex SVs, and SVs in repetitive regions, as long reads (> 5 kbp) can easily span repetitive sequences or SV breakpoints, and aligned precisely [22]. In the current study, the reads spanning the 11.2-kbp inversion in CFTR showed that the enhanced read length enables a full capture of SVs, significantly improving cancer SVs detection efficacy, providing a powerful tool for cancer precision therapeutics.
Gene fusions resulting from genomic rearrangements, represent an important part of tumor genomic landscape and are involved in development of approximately 16% of all cancer types, including CRC [39]. Although short-reads based whole genome sequencing (WGS) and RNA-seq are two major methods for identifying fusion genes, WGS is limited by the disadvantages mentioned above, and RNA-seq suffers from poor sensitivity for detecting the fusion genes that are expressed at rather low levels or diluted by accompanying non-cancerous cells [40]. In contrast, the advantages of long-read sequencing allow more effective identification of novel genetic rearrangements that may result in gene fusions. Indeed, our work uncovered a novel gene fusion, RNF38-RAD51B, which could enhance CRC cells’ oncogenic functions. RNF38 was reported as a vital driver of cancer progression and could promote the invasion and metastasis of cancer cells [30,31]. The RNF38-RAD51B gene fusion may enhance the expression or function of RNF38, since it significantly promoted the invasion and metastasis ability of colorectal cancer cells. Although the molecular mechanisms and clinical relevance of this gene fusion need to be further studied, our results suggest that nanopore sequencing may serve as a new strategy for detecting oncogenic gene fusions.
Nevertheless, this study has some limitations. First, the sample size (21 pairs of samples) was limited, making it difficult to find low-frequency somatic SVs in CRC. Second, a higher sequencing depth would be needed to improve the accuracy of SV phasing, especially for small insertions and deletions. Third, functional studies were required for further revealing functional roles of our newly-discovered somatic SVs, even though they were likely to promote development and progression of CRC according to their impact on genes structures (i.e., the inversions altered tumor suppressors APC and CFTR).
In summary, our study provides an example illustrating the utility of long-read nanopore sequencing in cancer genome investigation. Our work highlights the potential of the long-read sequencing in serving as a new platform for the precise diagnosis and treatment of CRC, and portrayed the first landscape of somatic SVs detected by long-read sequencing in CRC, which can be a useful resource for future biological and clinical studies.
Supporting information
The X-axis represents the patient IDs (detailed information see S1 and S2 Tables)
(PDF)
(A) The number of detected somatic SVs in MSS and MSI-H samples. (B) The number of detected somatic SVs in different stages.
(PDF)
(A and B) Quantification (A) and percentages of types (B) of somatic SVs detected by long-read sequencing in each sample. Insertions were the dominated SVs in MSIH samples. (C and D) Quantification (C) and percentages of types (D) of somatic SVs detected by long-read sequencing in each sample after the exclusion of insertions in STR regions. The X-axes in each graph represent the sample IDs.
(PDF)
(PDF)
The length (A) and distance (B) distributions of somatic SVs.
(PDF)
Quantification of singleton and recurrent somatic SVs in each sample including (A) or excluding (B) insertions in STR. The X-axes in each graph represent the patient IDs.
(PDF)
Different colors represent different recurrence number (left of the graph) within the tested tumor samples from 21 patients.
(PDF)
The X-axis represents the sample IDs.
(PDF)
(A) The 4,915 kbp inversion that affected APC in the sample C546-T. (B) The 11.2 kbp inversion that affected CFTR in the sample C564-T. (C) The RNF38-RAD41B gene fusion. (D) The SMAD3-SHISA6 gene fusion.
(PDF)
(A) The 4,915 kbp inversion that affected APC in the sample C546-T. (B) The 11.2 kbp inversion that affected CFTR in the sample C564-T.
(PDF)
The Sanger sequencing chromatograms of the breakpoints of the RNF38-RAD51B (A) and SMAD3-SHISA6 gene fusions (B).
(PDF)
The fusion gene was labelled by Flag tag.
(PDF)
(PDF)
(PDF)
(PDF)
(XLSX)
Data Availability
The sequence data were deposited in the Genome Sequence Archive (GSA) in the China National Center for Bioinformation (CNCB), under accession number HRA002638, that are publicly accessible (https://ngdc.cncb.ac.cn/gsa-human/browse/HRA002638).
Funding Statement
This work was supported by the National Natural Science Foundation of China (81773104, 81773263 to LW and 81873931, 81974382 to ZW), the Joint Fund of Ministry of Education for Equipment Pre-research (6141A02022626 to LW), the Major Scientific and Technological Innovation Projects in Hubei Province (2018ACA136 to ZW), the Integrated Innovative Team for Major Human Diseases Program of Tongji Medical College of HUST (to ZW), the Academic Doctor Supporting Program of Tongji Medical College, HUST (to ZW), and Health Commission of Hubei Province scientific research project (WJ2019M155 to ZW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. Epub 2018/09/13. doi: 10.3322/caac.21492 . [DOI] [PubMed] [Google Scholar]
- 2.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7(4):233–45. Epub 2007/03/16. doi: 10.1038/nrc2091 . [DOI] [PubMed] [Google Scholar]
- 3.Weischenfeldt J, Symmons O, Spitz F, Korbel JO. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet. 2013;14(2):125–38. Epub 2013/01/19. doi: 10.1038/nrg3373 . [DOI] [PubMed] [Google Scholar]
- 4.Yang R, Chen B, Pfutze K, Buch S, Steinke V, Holinski-Feder E, et al. Genome-wide analysis associates familial colorectal cancer with increases in copy number variations and a rare structural variation at 12p12.3. Carcinogenesis. 2014;35(2):315–23. Epub 2013/10/16. doi: 10.1093/carcin/bgt344 . [DOI] [PubMed] [Google Scholar]
- 5.Inaki K, Liu ET. Structural mutations in cancer: mechanistic and functional insights. Trends Genet. 2012;28(11):550–9. Epub 2012/08/21. doi: 10.1016/j.tig.2012.07.002 . [DOI] [PubMed] [Google Scholar]
- 6.Lee JA, Carvalho CM, Lupski JR. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell. 2007;131(7):1235–47. Epub 2007/12/28. doi: 10.1016/j.cell.2007.11.037 . [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Yang L, Kucherlapati M, Chen F, Hadjipanayis A, Pantazi A, et al. A Pan-Cancer Compendium of Genes Deregulated by Somatic Genomic Rearrangement across More Than 1,400 Cases. Cell Rep. 2018;24(2):515–27. Epub 2018/07/12. doi: 10.1016/j.celrep.2018.06.025 ; PubMed Central PMCID: PMC6092947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. Epub 2012/07/20. doi: 10.1038/nature11252 ; PubMed Central PMCID: PMC3401966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488(7413):660–4. Epub 2012/08/17. doi: 10.1038/nature11282 ; PubMed Central PMCID: PMC3690621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Audano PA, Sulovari A, Graves-Lindsay TA, Cantsilieris S, Sorensen M, Welch AE, et al. Characterizing the Major Structural Variant Alleles of the Human Genome. Cell. 2019;176(3):663–75 e19. Epub 2019/01/22. doi: 10.1016/j.cell.2018.12.019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nattestad M, Goodwin S, Ng K, Baslan T, Sedlazeck FJ, Rescheneder P, et al. Complex rearrangements and oncogene amplifications revealed by long-read DNA and RNA sequencing of a breast cancer cell line. Genome Res. 2018;28(8):1126–35. Epub 2018/06/30. doi: 10.1101/gr.231100.117 ; PubMed Central PMCID: PMC6071638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon JR, Xu J, Dileep V, Zhan Y, Song F, Le VT, et al. Integrative detection and analysis of structural variation in cancer genomes. Nat Genet. 2018;50(10):1388–98. Epub 2018/09/12. doi: 10.1038/s41588-018-0195-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helman E, Lawrence MS, Stewart C, Sougnez C, Getz G, Meyerson M. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24(7):1053–63. Epub 2014/05/16. doi: 10.1101/gr.163659.113 ; PubMed Central PMCID: PMC4079962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tubio JM. Somatic structural variation and cancer. Brief Funct Genomics. 2015;14(5):339–51. Epub 2015/04/24. doi: 10.1093/bfgp/elv016 . [DOI] [PubMed] [Google Scholar]
- 15.Jain M, Koren S, Miga KH, Quick J, Rand AC, Sasani TA, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 2018;36(4):338–45. Epub 2018/02/13. doi: 10.1038/nbt.4060 ; PubMed Central PMCID: PMC5889714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logsdon GA, Vollger MR, Eichler EE. Long-read human genome sequencing and its applications. Nat Rev Genet. 2020;21(10):597–614. Epub 2020/06/05. doi: 10.1038/s41576-020-0236-x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Zhao Y, Bollas A, Wang Y, Au KF. Nanopore sequencing technology, bioinformatics and applications. Nat Biotechnol. 2021;39(11):1348–65. Epub 20211108. doi: 10.1038/s41587-021-01108-x ; PubMed Central PMCID: PMC8988251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng S, Zhang MY, Wang XJ, Hu ZM, Li JC, Li N, et al. Long-read sequencing identified intronic repeat expansions in SAMD12 from Chinese pedigrees affected with familial cortical myoclonic tremor with epilepsy. J Med Genet. 2019;56(4):265–70. Epub 2018/09/09. doi: 10.1136/jmedgenet-2018-105484 . [DOI] [PubMed] [Google Scholar]
- 19.Aneichyk T, Hendriks WT, Yadav R, Shin D, Gao D, Vaine CA, et al. Dissecting the Causal Mechanism of X-Linked Dystonia-Parkinsonism by Integrating Genome and Transcriptome Assembly. Cell. 2018;172(5):897-909.e21. doi: 10.1016/j.cell.2018.02.011 ; PubMed Central PMCID: PMC5831509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bull RA, Adikari TN, Ferguson JM, Hammond JM, Stevanovski I, Beukers AG, et al. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat Commun. 2020;11(1):6272. doi: 10.1038/s41467-020-20075-6 ; Pubmed Central PMCID: PMC7726558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakamoto Y, Xu L, Seki M, Yokoyama TT, Kasahara M, Kashima Y, et al. Long read sequencing reveals a novel class of structural aberrations in cancers: identification and characterization of cancerous local amplifications. bioRxiv. 2019:620047. doi: 10.1101/620047 [DOI] [Google Scholar]
- 22.Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods. 2018;15(6):461–8. Epub 2018/05/02. doi: 10.1038/s41592-018-0001-7 ; PubMed Central PMCID: PMC5990442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Coster W, De Rijk P, De Roeck A, De Pooter T, D’Hert S, Strazisar M, et al. Structural variants identified by Oxford Nanopore PromethION sequencing of the human genome. Genome Res. 2019;29(7):1178–87. Epub 2019/06/13. doi: 10.1101/gr.244939.118 ; PubMed Central PMCID: PMC6633254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alaei-Mahabadi B, Bhadury J, Karlsson JW, Nilsson JA, Larsson E. Global analysis of somatic structural genomic alterations and their impact on gene expression in diverse human cancers. Proc Natl Acad Sci U S A. 2016;113(48):13768–73. doi: 10.1073/pnas.1606220113 ; Pubmed Central PMCID: PMC5137778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43(10):964–8. Epub 2011/09/06. doi: 10.1038/ng.936 ; PubMed Central PMCID: PMC3802528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi L, Guo Y, Dong C, Huddleston J, Yang H, Han X, et al. Long-read sequencing and de novo assembly of a Chinese genome. Nat Commun. 2016;7:12065. Epub 2016/07/01. doi: 10.1038/ncomms12065 ; PubMed Central PMCID: PMC4931320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burns KH. Transposable elements in cancer. Nat Rev Cancer. 2017;17(7):415–24. doi: 10.1038/nrc.2017.35 . [DOI] [PubMed] [Google Scholar]
- 28.Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, 3rd, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337(6097):967–71. Epub 2012/06/30. doi: 10.1126/science.1222077 ; PubMed Central PMCID: PMC3656569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cretu Stancu M, van Roosmalen MJ, Renkens I, Nieboer MM, Middelkamp S, de Ligt J, et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat Commun. 2017;8(1):1326. Epub 20171106. doi: 10.1038/s41467-017-01343-4 ; PubMed Central PMCID: PMC5673902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng R, Zhang PF, Yang X, Wei CY, Huang XY, Cai JB, et al. Overexpression of RNF38 facilitates TGF-β signaling by Ubiquitinating and degrading AHNAK in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38(1):113. Epub 2019/03/07. doi: 10.1186/s13046-019-1113-3 ; PubMed Central PMCID: PMC6402116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong D, Zhu SQ, Wu YB, Jin C, Jiang JH, Liao YF, et al. Ring finger protein 38 promote non-small cell lung cancer progression by endowing cell EMT phenotype. J Cancer. 2018;9(5):841–50. Epub 2018/03/28. doi: 10.7150/jca.23138 ; PubMed Central PMCID: PMC5868148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Z, Yang P, Ge H, Yang C, Cai Y, Chen Z, et al. RING Finger Protein 38 Mediates LIM Domain Binding 1 Degradation and Regulates Cell Growth in Colorectal Cancer. Onco Targets Ther. 2020;13:371–9. Epub 2020/02/06. doi: 10.2147/OTT.S234828 ; PubMed Central PMCID: PMC6969705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fleming NI, Jorissen RN, Mouradov D, Christie M, Sakthianandeswaren A, Palmieri M, et al. SMAD2, SMAD3 and SMAD4 Mutations in Colorectal Cancer. Cancer Res. 2013;73(2):725–35. doi: 10.1158/0008-5472.CAN-12-2706 . [DOI] [PubMed] [Google Scholar]
- 34.Yi K, Ju YS. Patterns and mechanisms of structural variations in human cancer. Exp Mol Med. 2018;50(8):98. Epub 2018/08/10. doi: 10.1038/s12276-018-0112-3 ; PubMed Central PMCID: PMC6082854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaisson MJ, Huddleston J, Dennis MY, Sudmant PH, Malig M, Hormozdiari F, et al. Resolving the complexity of the human genome using single-molecule sequencing. Nature. 2015;517(7536):608–11. Epub 2014/11/11. doi: 10.1038/nature13907 ; PubMed Central PMCID: PMC4317254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17(3):251–64. Epub 2015/03/25. doi: 10.1016/j.jmoldx.2014.12.006 ; PubMed Central PMCID: PMC5808190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, et al. Tumor Mutational Burden as an Independent Predictor of Response to Immunotherapy in Diverse Cancers. Mol Cancer Ther. 2017;16(11):2598–608. doi: 10.1158/1535-7163.MCT-17-0386 ; PubMed Central PMCID: PMC5670009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macintyre G, Ylstra B, Brenton JD. Sequencing Structural Variants in Cancer for Precision Therapeutics. Trends Genet. 2016;32(9):530–42. doi: 10.1016/j.tig.2016.07.002 . [DOI] [PubMed] [Google Scholar]
- 39.Valeri N. Streamlining Detection of Fusion Genes in Colorectal Cancer: Having "Faith" in Precision Oncology in the (Tissue) "Agnostic" Era. Cancer Res. 2019;79(6):1041–3. Epub 2019/03/17. doi: 10.1158/0008-5472.CAN-19-0305 . [DOI] [PubMed] [Google Scholar]
- 40.Heyer EE, Deveson IW, Wooi D, Selinger CI, Lyons RJ, Hayes VM, et al. Diagnosis of fusion genes using targeted RNA sequencing. Nat Commun. 2019;10(1):1388. Epub 2019/03/29. doi: 10.1038/s41467-019-09374-9 ; PubMed Central PMCID: PMC6437215 [DOI] [PMC free article] [PubMed] [Google Scholar]