Figure 1: HAQERs, the fastest-evolved regions of the human genome.
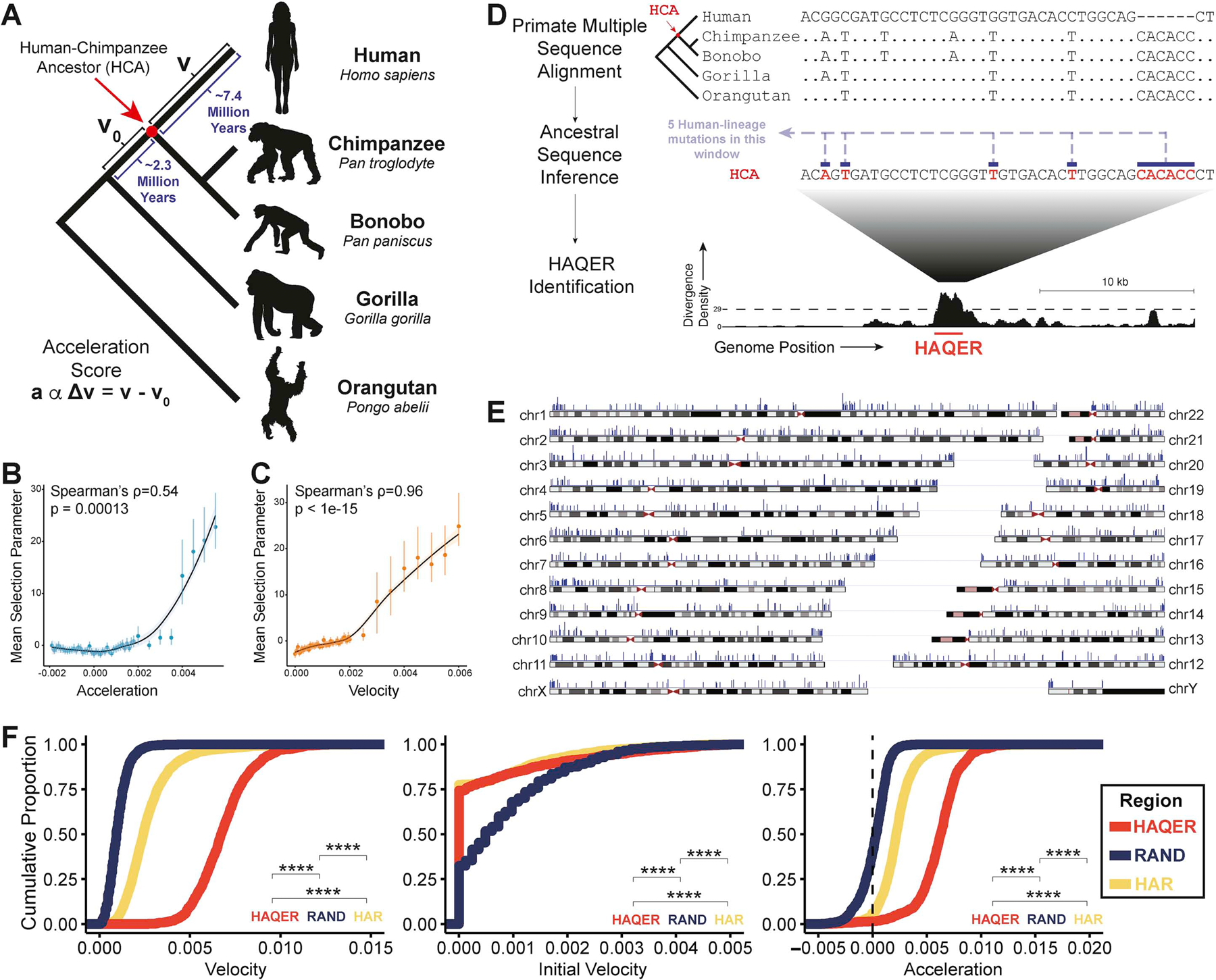
(A) We display the values of velocity (v), initial velocity (v0), and acceleration (a) in the phylogenetic context of recent human evolution. (B, C) Mean selection parameter estimates for 500bp genomic regions binned by either acceleration (B) or velocity (C). Error bars display the 95% highest density credible interval. Both acceleration and velocity correlate with signatures of selection in human populations. (D) HAQERs (Human Ancestor Quickly Evolved Regions) are identified as regions containing at least 29 mutations in a 500bp window (p < 10−6) that separate the inferred human-chimpanzee ancestor sequence from the human genome. We count insertions and deletions as one mutation regardless of their length. (E) Locations in the human genome of the 1581 HAQERs (blue markers). Marker amplitude reflects the maximum divergence density observed in each region. HAQERs are distributed across all human chromosomes and enriched near chromosome ends. (F) Cumulative distribution of velocity, initial velocity, and acceleration observed across HAQERs, Human Accelerated Regions (HARs), and random neutral proxy regions (RAND). Regions are filtered to a minimum element size of 50bp. (Bonferroni-adjusted Wilcoxon, ****; p < 0.0001). See also Figures S1–2.
