Figure 4: Rapid sequence divergence in HAQERs generated hominin-specific neurodevelopmental enhancers.
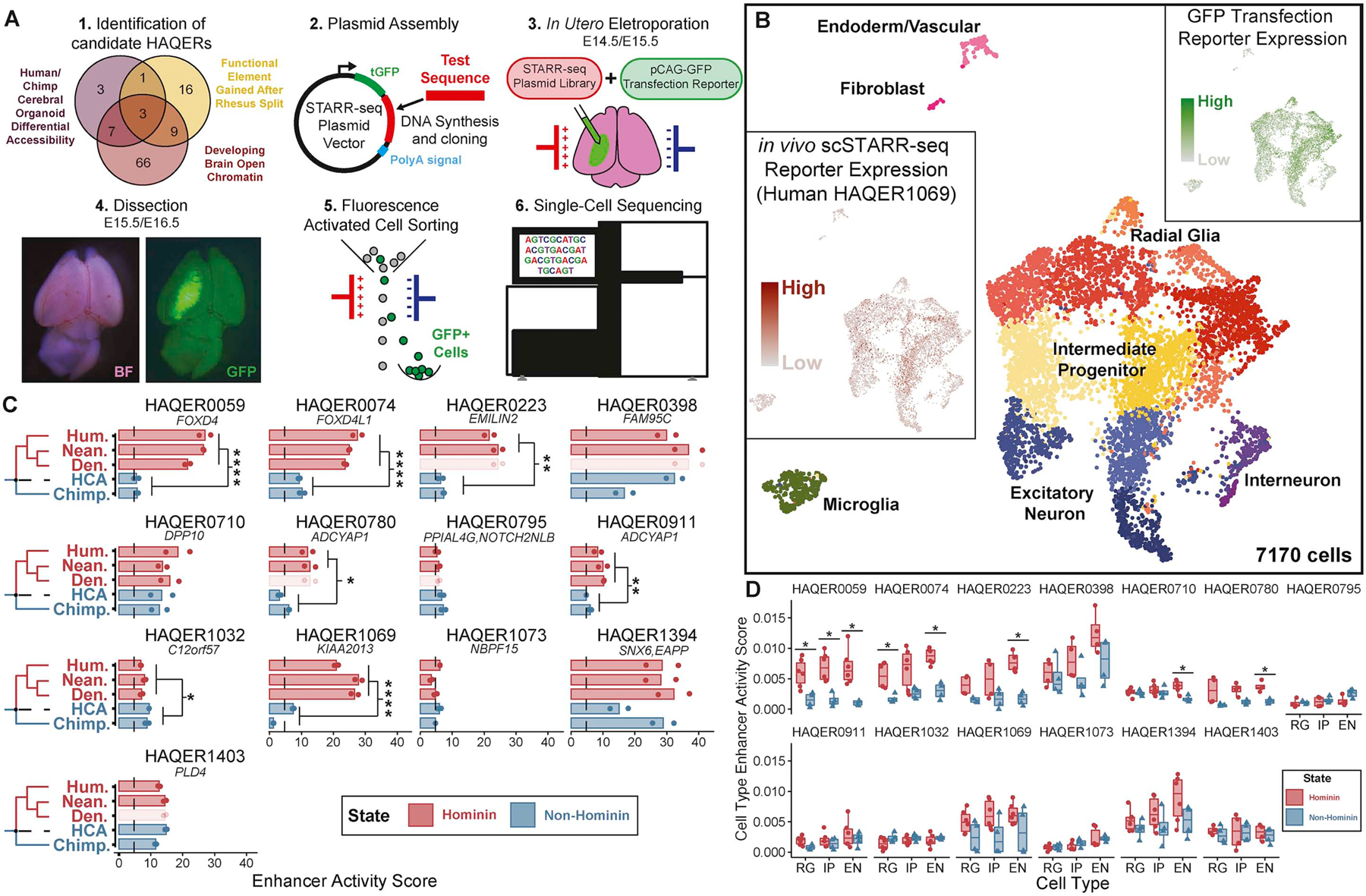
(A) Experimental design. Candidate HAQERs were prioritized based on overlaps with epigenomic datasets, cloned into a STARR-seq vector, and electroporated into developing mouse brains along with a pCAG-GFP transfection reporter. Single-cell sequencing followed dissection and FACS enrichment of GFP+ cells. (B) UMAP projection of 7,170 single cells from two scSTARR-seq experiments, labeled with metacluster identities. Inserts display cells colored by expression of the GFP transfection reporter and Human HAQER0169. (C) Enhancer activity score, defined as the input-normalized UMI count pooled across all cells per 1000 reporter UMIs, for 13 HAQERs. Nearest gene name is displayed below each HAQER ID. We display significant differences in enhancer activity between Hominin (Human, Neanderthal, Denisova) and non-Hominin (Chimpanzee, human-chimpanzee ancestor (HCA)) sequences (Bonferroni-adjusted t-test, p < 0.05). Faded bars represent sequences where Neanderthal and Denisovan had the same sequence in the 500bp genomic region and these duplicate sequences are not included in the statistical analysis. (D) Cell type enhancer activity score, or the input-normalized reporter UMI count normalized to the pCAG-GFP UMI count for each cell averaged across all cells in each metacluster (RG=Radial Glia, IP=Intermediate Progenitor, EN=Excitatory Neuron) (FDR- corrected t-test, p < 0.05). See also Figure S5–6.
